Spatiotemporal transcriptomic atlas of planarian
Summary
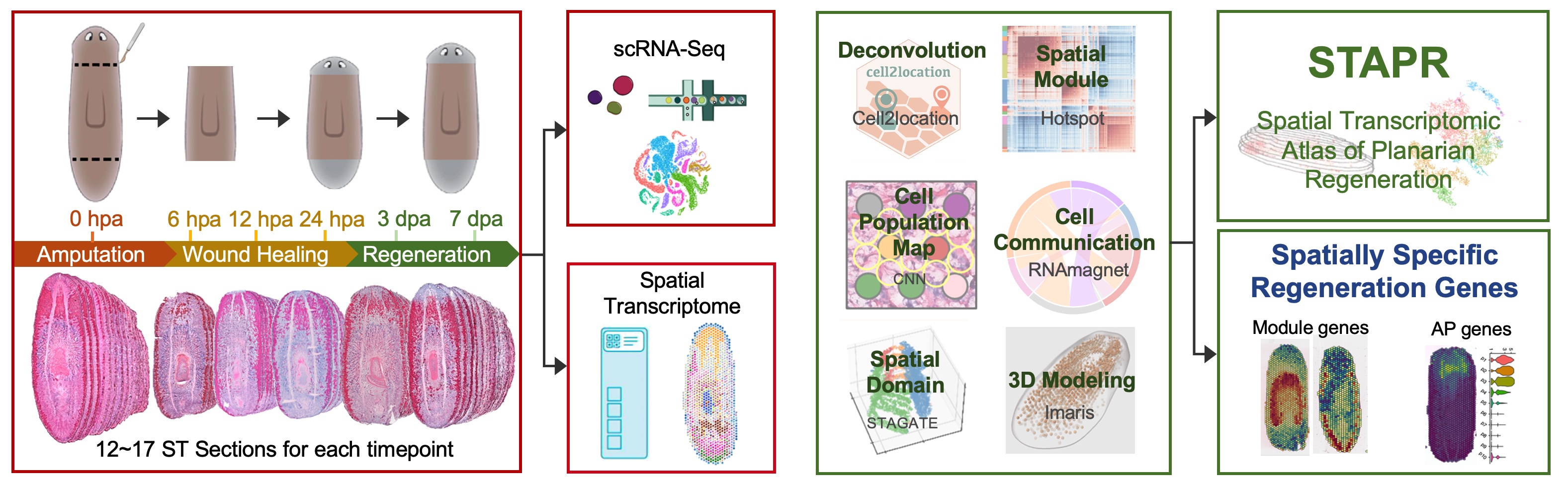
We generated the spatiotemporal transcriptomic atlas of planarians across six time points of regeneration process using 10x Visium technology. The samples include 17 whole-worm coronal cryosections at 0 hours post-amputation (hpa), 17 at 6 hpa, 12 at 12 hpa, 12 at 24 hpa, 15 at 3 days post-amputation (dpa), and 16 at 7 dpa. Meanwhile, we generated a set of single-cell transcriptomic data from the same batch of samples collected from the identical time points as ST plus 2 dpa and 5 dpa time points. In total, we collected transcription profiles of 59,506 spatial spots and 55,014 single cells from the regeneration process. To reconstruct 3D cell distribution profiles of planarians at each regenerating time point, we took advantage of the latest data analysis tools such as Cell2location, Hotspot, STAGATE and Imaris.
Species
Schmidtea mediterranea
Technology
10x Chromium and 10x Visium
Stages
Spatial Transcriptome: 0 hpa, 6 hpa, 12 hpa, 24 hpa, 3 dpa, 7 dpa
Single Cell: 0 hpa, 6 hpa, 12 hpa, 24 hpa, 2 dpa, 3 dpa, 5 dpa, 7 dpa