circAtlas 3.0 a gateway to 3 million standardized named vertebrate circular RNAs
How to use
Introduction
Welcome to circAtlas (An integrated resource of circRNAs in vertebrates).
circAtlas contains a compendium of 1,070 RNA-seq libraries from 19 normal tissues from human, macaca, mouse, rat, pig and chicken.
Main characteristics of circAtlas include:
circAtlas introduces an ID assignment scheme,by integrating sequence conservation and expression patterns of circRNAs.
Search and find conserved circRNAs across human, macaca, mouse, rat, pig, chicken.
Convert the circRNA ID among different circRNA databases.
Predict potential peptide-encoding circRNAs. e.g: ORF, IRES.
Predict regulation elements of circRNAs. e.g: miRNA, RBP.
circAtlas employs three types of networks including gene co-expression, miRNA regulation and RBP regulation to infer circRNA functions.
Data
Users can select one of the six species or input cirRNA IDs to browse circRNAs, which were identified by CIRI2, CIRI-full, DCC, find_circ and CIRCexplorer2.

| Columns | Description |
|---|---|
| MCS | 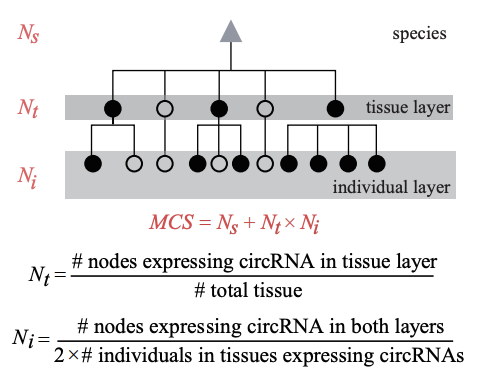 |
| TIS | Tissue specifity index. |
Users can get the detail information of each circRNA when click the circAltas ID.

Search circAtlas
1.Users can search the circRNA known in circAtlas via circAltas ID, chromosome position, host gene and the name in other circRNA database.
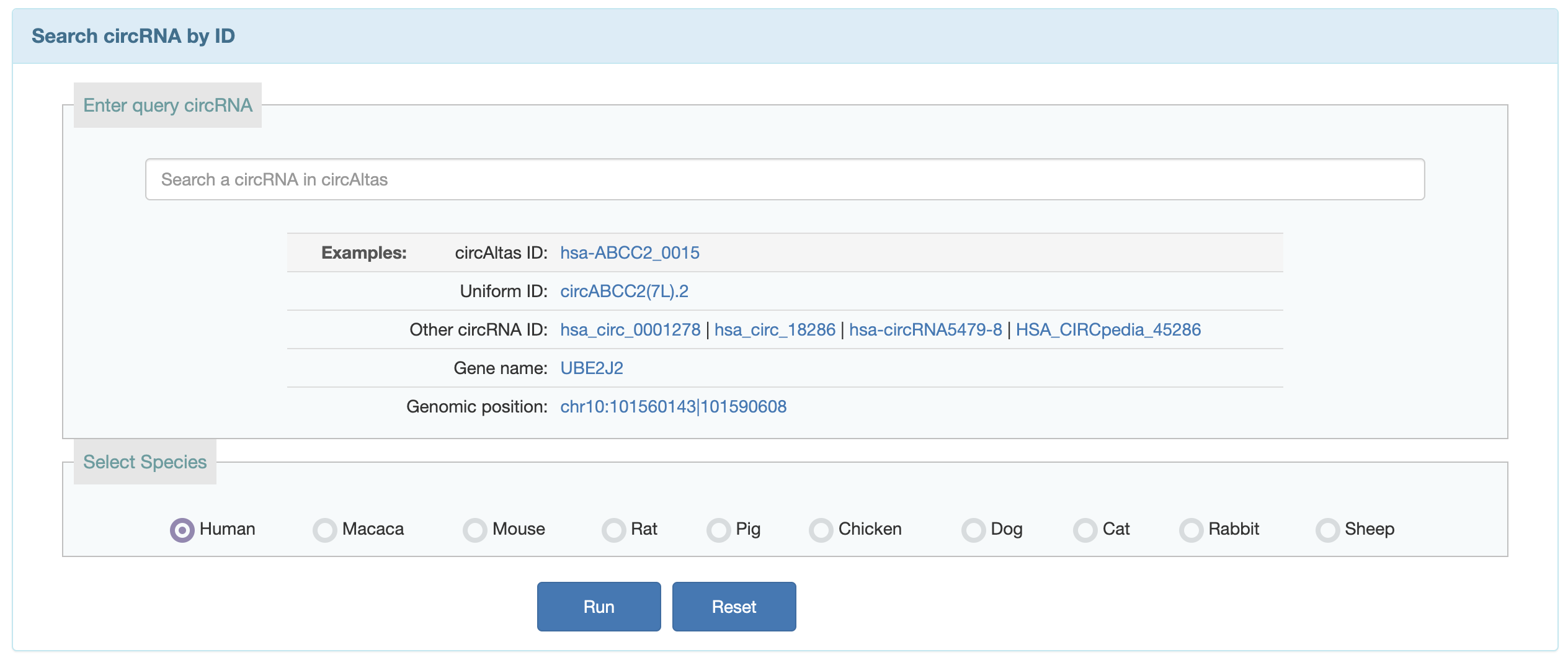
2.Users can search the circRNA known in circAtlas which have potential function, such as:
- Conversed circRNAs
- ORF
- IRES
- RBP
- miRNA
- Network annotation

Analyze
Conserved circRNAs
User can search conserved circRNAs on currently available data or predict conserved circRNAs from a corresponding input.
Input:- Input your list of query circRNAs into the first box in the panel.
- ISelecct the species of query circRNAs.
- Choose additional tag options:
- Min similarity: Sets minimum sequence identity (in percent). Default is 60 for circRNAs searches.
- Submit the request by clicking Run button.

Output: After the analysis has finished you will end up at an overview table displaying all the detected conserved circRNAs.

ORF
User can search predicted IREs on currently available circRNAs or predict IREs on given circRNAs from a corresponding input.
Input:- Input your list of query circRNAs in FASTA format.The FASTA format includes: a header line that starts with '>' and followed by a line containing a sequence.
- Choose additional tag options:
- Min ORF length: Sets minimum sequence identity (in percent). Default is 60 for circRNAs searches.
- Output type: Sets minimum score. This is the matches minus the mismatches minus some sort of gap penalty. Default is 60.
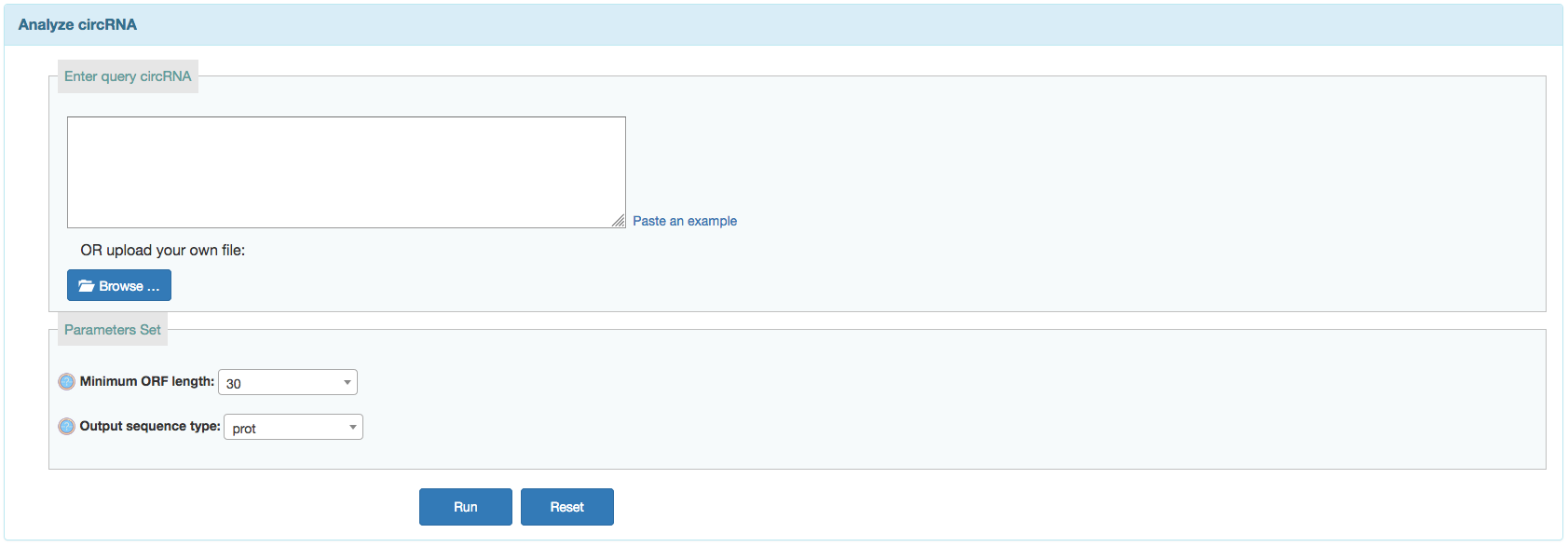
Output: After the analysis has finished you will end up at an overview table displaying all the detected ORFs on circRNAs. Users can click the row in table to get the protein and nucleotide acid sequence.
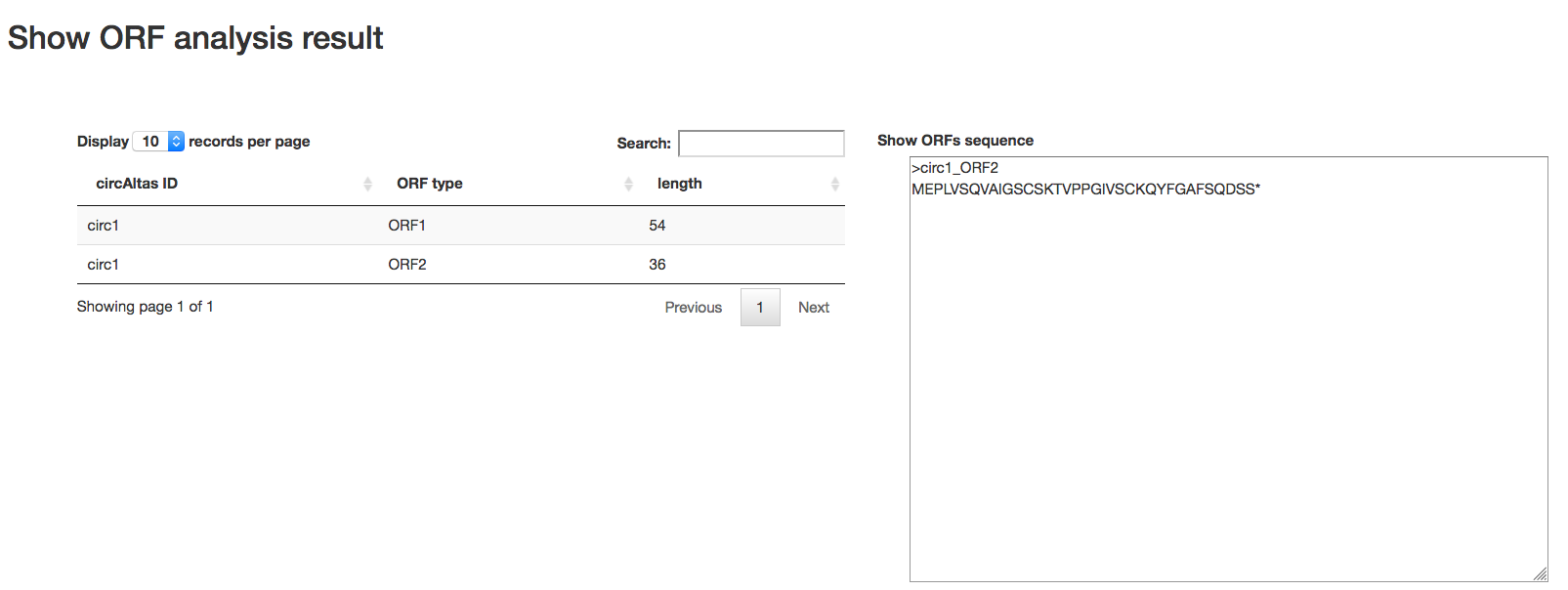
IRES
User can search predicted IREs on currently available circRNAs or predict IREs on given circRNAs from a corresponding input.
Input:- Input your list of query circRNAs in FASTA format.The FASTA format includes: a header line that starts with '>' and followed by a line containing a sequence.
- Choose additional tag options:
- Handel Model:
- 0,default, just using the complete transcript sequences;
- 1, optional, using the subsequences specified by the index (e.g. >id start_pos end_pos);
- 2, optional, spliting the transcript into fragments, please provides the window size (-w, defualt 174) and the step size (-s, defualt 50);
- Sliding window size: Ignore this option if the selected model is not 2. Default is 174.
- Sliding window step: Ignore this option if the selected model is not 2. Default is 50.
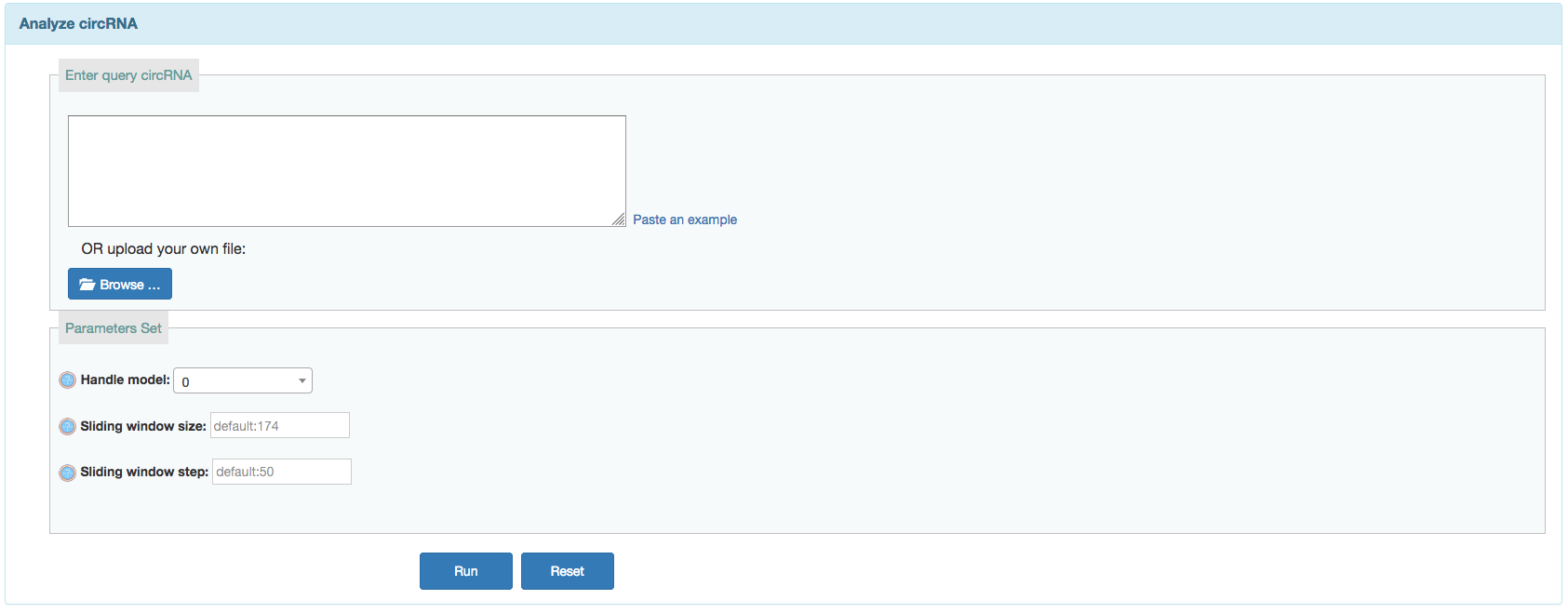
Output: After the analysis has finished you will end up at an overview table displaying all the detected ORFs on circRNAs. Users can click the row in table to get the sequence.

RBP
User can search predicted RBP binding on currently available circRNAs or predict RBP binding sites on given circRNAs from corresponding input.
Input:- Input your list of query circRNAs in corresponding format. Users want predicted the RBP binding sites in circRNA sequence must input the full sequence.
- Choose additional tag options:
- Region: Predict RBP bingding sites in upstream, downstream, and sequence.
- RNA-binding Proteins: The RNA-binding Protein all from POSTAR2 and starBase databases.Users can choose all factor or select several factors.
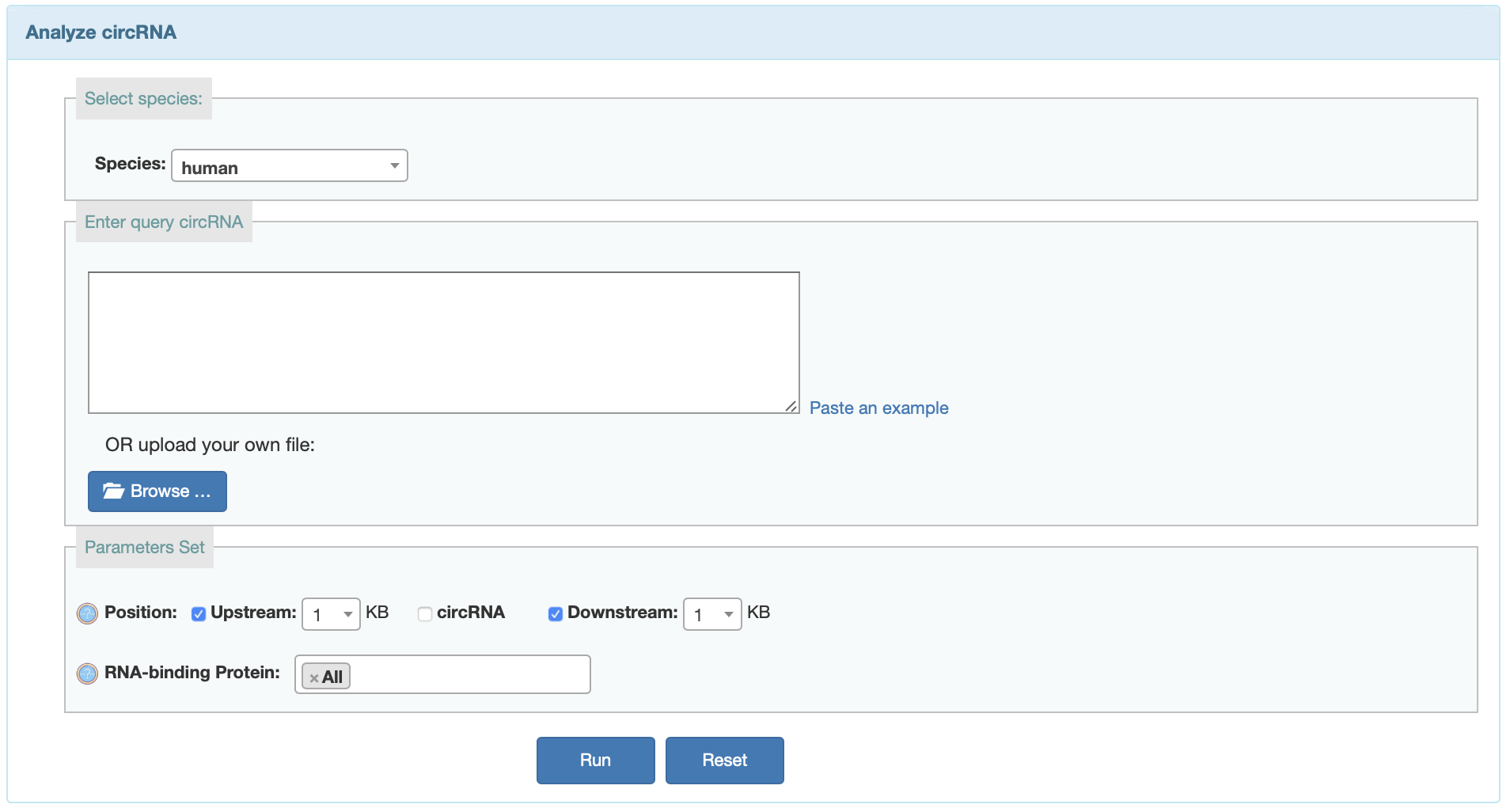
Output: After the analysis has finished you will end up at an overview table displaying all the detected RBP binding sites on circRNAs.
miRNA
User can search predicted miRNA binding on currently available circRNAs or predict miRNA binding sites on given circRNAs from acorresponding input.
Input:- Input your list of query circRNAs in FASTA format.The FASTA format includes: a header line that starts with '>' and followed by a line containing a sequence.
- Choose additional tag options:
- Software:Users can choose different software to predict the miRNA binding sites. Pita will take several minutes.

Output: After the analysis has finished you will end up at an overview table displaying all the detected miRNA binding sites on circRNAs.
Network
User can search functional annotation of currently available circRNAs or annotate given circRNAs by constructing network.
Sample data: Clicking on the 'example' button downloads an example of an input list.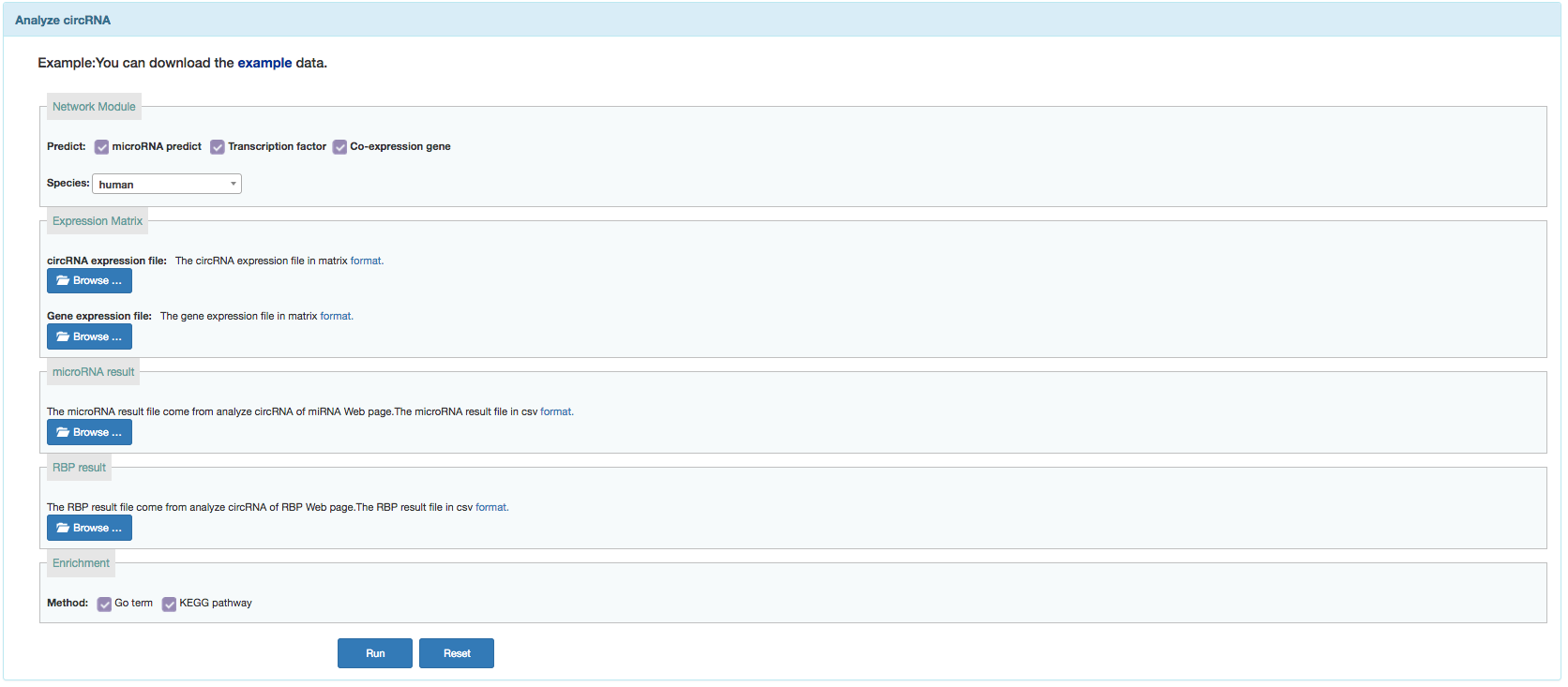
Input:
- Input query circRNAs file in corresponding format.
- circRNA expression matrix:in matrix format.
- gene expression matrix:in matrix format.
- miRNA result:in csv format.Download the result from miRNA result page.
- RPB result:in csv format.Download the result from RBP result page.
- Choose additional tag options:
- Annotation method:GO term and KEGG pathway.
Disease
The page provides disease related circRNA by integrating three databases: circad, CircR2Disease and circRNADisease.
With this module, users can do searches and queries on target circRNAs.
Users can also submit new records by providing your work email address, where you will receive the confirmation request. If there are any problems with your future submissions, the circAtlas curators will contact you at this email address. Please note that your email address will be presented on the web in a way that should protect it from spammers.