Date, November 16, 2015
To: Canine Genomics Community
From: Ya-‐ping Zhang, Ph.D.
Robert Wayne, Ph.D.
Elaine A. Ostrander, Ph.D.
Invitation Letter
Dear Colleagues,
We are developing a consortium to undertake a dramatic complete genome sequencing effort in dogs and gray wolves. We already secured funding for the first stage of 1000 complete genomes (see below) and would like to have your advice and expertise as part of the group archiving and analyzing the sequence date. Some of our specific questions are oriented toward understanding the evolutionary history of dogs, genetic changes associated with domestication and relationships of genetic changes to phenotype. We also have a goal to investigate loci involved in canine susceptibility to disease, particularly among certain breeds. Our final interest is in understanding the genetic variation between breeds using whole genome sequence data. The initiative will be co-‐led by Ya-‐ping Zhang, Robert Wayne, and Elaine Ostrander.
The aim of the Dog10k is to coordinate the global effort on genome sequencing in dogs and build a comprehensive resource for the canine community (similar to the 1000 genome project for humans). We envision a 5-‐year project with three stages as follows:
In stage one, our initial plan is to undertake whole genome sequencing of ~1000 dogs. Two hundred of these will be sequenced at high coverage (~30X) intended for detailed demographic inference and 800 will be sequenced with lower coverage sequencing (~15X), further supporting the evolutionary analysis. We are hoping to achieve the first stage goals by the end of 2016. This goal may change based on the number and quality of whole genome sequence data already available, or about to be made available, by the community.
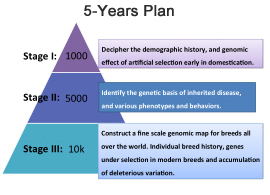
In order to facilitate coordination and collaboration, we intended to hold a conference in May 2016 in Beijing, China. At the meeting, we plan to discuss preliminary results from analysis of over ~500 dogs and wolves from whom at least partial data are available. We will develop specific mechanisms for collaboration, and data and sample sharing. Consideration of how the sequencing effort should be divided between breed dogs, wild dogs, and wild canids is a major a
critical point. Also, since a major goal is to develop a resource for disease gene mapping, number of controls that should be sequenced at high or low density for breeds affected with disorders of interest will need to be considered as well.
We have secured funding to provide travel and accommodation for 15 key speakers (oral presentation) and attendees, and are hoping you might honor us with attending and potentially being part of the consortium. Please respond direct to any of the organizers, and we hope to see you in Beijing!
Yours sincerely,
Ya-‐Ping Zhang
Robert Wayne
Elaine A. Ostrander