Difference between revisions of "ENST00000602812.1"
Chunlei Yu (talk | contribs) |
Chunlei Yu (talk | contribs) (→Transcriptomic Nomenclature) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | Ftx, a conserved functional long ncRNA,which located in the cis-acting regulatory region of the imprinted X-chromosome inactivation(XCI), is a positive regulator of Xist. | + | ''Ftx'', a conserved functional long ncRNA, which located in the cis-acting regulatory region of the imprinted X-chromosome inactivation(XCI), is a positive regulator of Xist. |
==Annotated Information== | ==Annotated Information== | ||
| − | === | + | ===Name=== |
| − | FTX, FTX transcript, XIST regulator (non-protein coding)(HGNC nomenclature) | + | ''FTX'', FTX transcript, XIST regulator (non-protein coding)(HGNC nomenclature) |
| + | |||
| + | "five prime to XIST", ''FLJ33139'', ''LINC00182'' <ref name="ref1" /> | ||
| − | |||
===Function=== | ===Function=== | ||
| − | [[File:A model for Ftx action on Xist.gif|right|thumb|400px'''A model for Ftx action on Xist.'''<ref name="ref1" />]] | + | [[File:A model for Ftx action on Xist.gif|right|thumb|400px|'''A model for Ftx action on Xist.'''<ref name="ref1" />]] |
| + | |||
Ftx is specifically upregulated during female ES cell differentiation at the onset of XCI. Loss of Ftx expression through the deletion of the promoter region leads to local alteration in the chromatin structure. More importantly, Ftx impacts on gene expression within theXic and in particular on Xist, as Xist is significantly downregulated in the absence of Ftx <ref name="ref1" />. | Ftx is specifically upregulated during female ES cell differentiation at the onset of XCI. Loss of Ftx expression through the deletion of the promoter region leads to local alteration in the chromatin structure. More importantly, Ftx impacts on gene expression within theXic and in particular on Xist, as Xist is significantly downregulated in the absence of Ftx <ref name="ref1" />. | ||
| − | Disruption of Ftx did not affect the survival of female embryos or expression of Xist and other X-linked genes in the preimplantation female embryos, which indicates that Ftx is dispensable for imprinted XCI in preimplantation embryos <ref name="ref2" /> | + | Disruption of Ftx did not affect the survival of female embryos or expression of Xist and other X-linked genes in the preimplantation female embryos, which indicates that Ftx is dispensable for imprinted XCI in preimplantation embryos <ref name="ref2" />. |
===Cellular Localization=== | ===Cellular Localization=== | ||
| − | [[File:Subcellular localization of Ftx transcripts.gif|right|thumb|400px'''Subcellular localization of Ftx transcripts.'''<ref name="ref1" />]] | + | [[File:Subcellular localization of Ftx transcripts.gif|right|thumb|400px|'''Subcellular localization of Ftx transcripts.'''<ref name="ref1" />]] |
| − | + | It produces a transcript with no apparent coding potential that predominantly remains in the nucleus <ref name="ref1" />. | |
===Expression=== | ===Expression=== | ||
| Line 20: | Line 22: | ||
===Evolution=== | ===Evolution=== | ||
| − | Ftx is well conserved during evolution and contains several microRNAs (miRs) which are also conserved <ref name="ref3" /> | + | Ftx is well conserved during evolution and contains several microRNAs (miRs) which are also conserved <ref name="ref3" />. |
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
| − | Unité de Génétique Moléculaire Murine, URA 2578, Institut Pasteur, Paris, France <ref name="ref1" /> | + | Unité de Génétique Moléculaire Murine, URA 2578, Institut Pasteur, Paris, France <ref name="ref1" />. |
Department of Epigenetics, Medical Research Institute, Tokyo Medical & Dental University, 1-5-45 Yushima Bunkyo-ku, Tokyo, 113-8510, Japan <ref name="ref2" />. | Department of Epigenetics, Medical Research Institute, Tokyo Medical & Dental University, 1-5-45 Yushima Bunkyo-ku, Tokyo, 113-8510, Japan <ref name="ref2" />. | ||
| − | Young Investigators Group Bioinformatics and Transcriptomics, Department Proteomics, Helmholtz Centre for Environmental Research - UFZ, Leipzig, Germany, and Department for Computer Science, University of Leipzig, Leipzig, Germany; RNomics Group, Department Diagnostics, Fraunhofer Institute for Cell Therapy and Immunology - IZI, Leipzig, Germany <ref name="ref3" /> | + | Young Investigators Group Bioinformatics and Transcriptomics, Department Proteomics, Helmholtz Centre for Environmental Research - UFZ, Leipzig, Germany, and Department for Computer Science, University of Leipzig, Leipzig, Germany; RNomics Group, Department Diagnostics, Fraunhofer Institute for Cell Therapy and Immunology - IZI, Leipzig, Germany <ref name="ref3" />. |
==References== | ==References== | ||
| Line 41: | Line 43: | ||
tID = ENST00000602812.1| | tID = ENST00000602812.1| | ||
source = Gencode19| | source = Gencode19| | ||
| − | same = NONHSAT137589| | + | same = NONHSAT137589,''FTX'',''LINC00182'', ''FLJ33139''| |
classification = intergenic| | classification = intergenic| | ||
length = 2356 nt| | length = 2356 nt| | ||
Latest revision as of 06:24, 25 June 2016
Ftx, a conserved functional long ncRNA, which located in the cis-acting regulatory region of the imprinted X-chromosome inactivation(XCI), is a positive regulator of Xist.
Contents
Annotated Information
Name
FTX, FTX transcript, XIST regulator (non-protein coding)(HGNC nomenclature)
"five prime to XIST", FLJ33139, LINC00182 [1]
Function
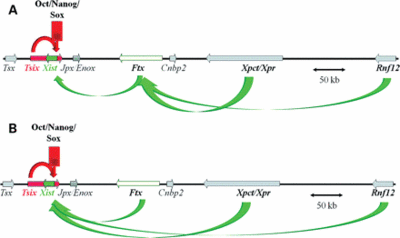
Ftx is specifically upregulated during female ES cell differentiation at the onset of XCI. Loss of Ftx expression through the deletion of the promoter region leads to local alteration in the chromatin structure. More importantly, Ftx impacts on gene expression within theXic and in particular on Xist, as Xist is significantly downregulated in the absence of Ftx [1]. Disruption of Ftx did not affect the survival of female embryos or expression of Xist and other X-linked genes in the preimplantation female embryos, which indicates that Ftx is dispensable for imprinted XCI in preimplantation embryos [2].
Cellular Localization
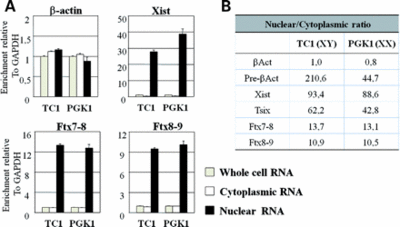
It produces a transcript with no apparent coding potential that predominantly remains in the nucleus [1].
Expression
FTX identified as chromatin-associated lncRNA was predominantly downregulated in breast tumor [3].
Evolution
Ftx is well conserved during evolution and contains several microRNAs (miRs) which are also conserved [3].
Labs working on this lncRNA
Unité de Génétique Moléculaire Murine, URA 2578, Institut Pasteur, Paris, France [1].
Department of Epigenetics, Medical Research Institute, Tokyo Medical & Dental University, 1-5-45 Yushima Bunkyo-ku, Tokyo, 113-8510, Japan [2].
Young Investigators Group Bioinformatics and Transcriptomics, Department Proteomics, Helmholtz Centre for Environmental Research - UFZ, Leipzig, Germany, and Department for Computer Science, University of Leipzig, Leipzig, Germany; RNomics Group, Department Diagnostics, Fraunhofer Institute for Cell Therapy and Immunology - IZI, Leipzig, Germany [3].
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Chureau C, Chantalat S, Romito A, Galvani A, Duret L, Avner P, et al. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region[J]. Human molecular genetics. 2011,20(4):705-18.
- ↑ 2.0 2.1 Soma M, Fujihara Y, Okabe M, Ishino F, Kobayashi S. Ftx is dispensable for imprinted X-chromosome inactivation in preimplantation mouse embryos[J]. Scientific reports. 2014,4:5181.
- ↑ 3.0 3.1 3.2 Soma M, Fujihara Y, Okabe M, Ishino F, Kobayashi S. Ftx is dispensable for imprinted X-chromosome inactivation in preimplantation mouse embryos[J]. Scientific reports. 2014,4:5181.
Basic Information
| Transcript ID |
ENST00000602812.1 |
| Source |
Gencode19 |
| Same with |
NONHSAT137589,FTX,LINC00182, FLJ33139 |
| Classification |
intergenic |
| Length |
2356 nt |
| Genomic location |
chrX-:73247971..73513409 |
| Exon number |
7 |
| Exons |
73247971..73249386,73251431..73251675,73494177..73494245,73500229..73500329,73500766..73500920,73501537..73501683,73513187..73513409 |
| Genome context |
|
| Sequence |
000001 GGAAGCACAC TGCGGCGATT CTGGAGAGGT TGTCCTTTTC TGGCATCAGC GTTGCGGCTC GAGGCAAAGC TGGTCCTGTG 000080
000081 CCTGCTGTCC ATTGCCGCTA AGGCCTGCTG CTGCCATTCA GGCGTGTCTC TCTCTCTCTC TCTCTTTCAT GCCGCTCTCT 000160 000161 CCACCCCATC TTGGGACGGC CAGCTCTAAG AATTCCTGCT ACGACACTGA ATTCAACCAG GAGGTGATGC CACACTCAAT 000240 000241 CAGAAACTAT GCTACTGCTG AAGAGGTATA TTGAGCCCTG CCTGGTTAAC CTCCAGTGAG GTTATTAGAG AATTGGGAGA 000320 000321 GGCTCAAATC AGCATCCTGC ACCTAGTTAT CAAACCATTG TGAAGTCTTG ATTCCTGGAT GGCCAGAAAA CTTGTCATTG 000400 000401 GTCAAGAGCA ACATTAAGAT CTGCCTGTTA CTCATACTGA TGCTTCCTGC CCAGCAAGTT CATCAGAGGG CGAGGATGAA 000480 000481 TTACGGTTGC CATGAATGTC CTTGTGAGGC AGTTGGGAAG CCTTGGCTGG ATTGTGAAGG ATATTTGAAA AGTATTTTAG 000560 000561 AGTCCACCAG ATCATCCATG TGAGTGACCA TGCAGCAAAA GAAGGAGGGT CCTCTCAGTG ATTAAGGATT ATGAAGTATT 000640 000641 AATTCTTCCA CTGATATCTC AAAAGGTGAT CTGGACAAAG GAACAGGTAT CTCAGGATTC CTCCTTGGTC ACTTTTCAGC 000720 000721 AGCAGCCTGC TCTTCTTTTG CAATCTCTTC AGGATCTCTG TAGAAGTAGA GATCAGGCAT GAACTCCCAC GGGTGTTCAC 000800 000801 GGGAAATGGT GCCACGCATG CGCAGAACTT CCCGAGCCAG CATCCACCAC ATCAAACCCA CTGAGTGAGC TCCCTTGTTG 000880 000881 TTGCATGGGA TGGCAATGTC CACGTATCAC AGAGGAGAAT CTGTGTTACA CAGAGCAATG TAGAGACGGG GTTTCACCGT 000960 000961 GTTAGCCAGG ATGGTCTCGA TCTCCTGACC TCATGATCCA CCCGCCTTGG CCTCCCAAAG TGCTGGGATT ACAGGCGTGA 001040 001041 ACCACCGCGC CCGGCCAGGA GAGTGTATTT TCTACACATA AGTTCAGGTC ATCTCCACAC AGACCCAGTT TGCCTCCCTC 001120 001121 TTTTCCGATC TGAATGCCTT TAATTTCCTT CTCTTGCCTA ACTGTACTGG CTAGAACATC CCGAACTAGG TTGAATGAGG 001200 001201 TGCTGAGAGT GGGCATAATT GGCTTCTTTA CCTCCGAAGA AAATATTTCA TGTTTTCTTC ATTGATTATG ATGTTAGTTG 001280 001281 TGGGTATTTC ATTTACTGCC TATATGGTAT AAAGATACAT TCCTTCTATG CCTAATTGGT TGACAGCTTT TATTATATCA 001360 001361 AATTCTATAA AACTGTTTTA TTTTTAAATC TATTGAGATG ATCAAATGGC GGTGTCCCTC CTCCTTTATA AGACTATTTA 001440 001441 AAACATCCTT GCCTCAGAGG GACAAATCCC ATTTGAACAT GGCAAGTAAT CCTCTTAATA TAAAGTGGAA TTAGGTTTGA 001520 001521 CAGAAATTAC CAGTGAAGCA CTATGGGCAT ATATTTGATA GGAGATTTTT ATTATGGATA CAATTTACTT GTTATTGGTC 001600 001601 TCTTCAGATA TTCTTTCTCA TGACTGGATT TGCGTACGTT GTATGAGTAC AAATTTTTTC ATTTATTCTA CGCTGCTCTT 001680 001681 TATTATATGC TTCCTTCTGT AAAATTTGGA CTGTTTGTTC TTCCCATTAT ATTAATAGTT GCTTCAGGTG TAACATAAGG 001760 001761 TTATTTTTTA AGTACTTTTC TGGTATCACA AATATTGGTG TTTTGTGTTT CATCTTTCAT TTGTCAAAAT ATTTTTTAAC 001840 001841 TTCCATTTGG ATTTAATGAT TCACCCATTC ATTATTCAAA AGCATATTGT TTAATTTCTA TGTGTTTGTG AACATTCTAA 001920 001921 TGTTCATTCT ACTAATAGTT CCTAGGTTCA TGCAATGTGC TTAAAAATGT GGTATAATTT CTGTATTATT AAATGTAAAA 002000 002001 CTAGTCTTGA TTCGTAACAT AAGGCCTACG CAAAGATCCA TCTTCTGCAG AGAAGAATGT AGATTGTGCC GCTGTTAGAC 002080 002081 GAAGTATTCT GTATACGTCA GTTTGCTTTA TTTGGCATAA AGTGTAGTGT AAGCCAGATG TAGTGTAAGT CAGATCCAGA 002160 002161 TGATCTGCCC ATTGCTGAAA CTTCAGTACT TAAGTCCCCT ATTATTATTG TATGGCAATC TATCTGTCCA CTCAGAAGTT 002240 002241 ATTATATTTG TTTTATATGA TTGAGCACTT CTGTTTTGGG TATATATGTA TTTACAATTA CTATAACCTC TTGCTGAATT 002320 002321 AACCCCTTTT ACCATTATGT AATAAACTTA TTTCCC |