Difference between revisions of "MAFTRR"
| (5 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
===Approved Name=== | ===Approved Name=== | ||
MAF transcriptional regulator RNA | MAF transcriptional regulator RNA | ||
| − | |||
| − | |||
===Synonyms=== | ===Synonyms=== | ||
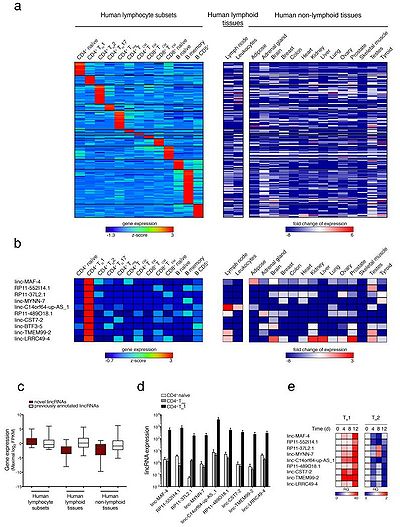
| + | [[File: MAFTRR.jpg|right|thumb|400px|'''Figure: LincRNA signatures of human lymphocyte subsets.''' <ref name="ref1" />.]] | ||
| + | |||
linc-MAF-4 | linc-MAF-4 | ||
===Chromosome=== | ===Chromosome=== | ||
| Line 34: | Line 34: | ||
===Diseases=== | ===Diseases=== | ||
| − | + | ||
* allergy | * allergy | ||
Latest revision as of 01:56, 30 June 2016
Annotated Information
Approved Symbol
MAFTRR
Approved Name
MAF transcriptional regulator RNA
Synonyms
linc-MAF-4
Chromosome
16q23.2
RefSeq ID
NR_104663
OMIM ID
616264
Ensembl ID
ENSG00000261390
pubmed IDs
25621826
Characteristics
linc-MAF-4,a TH1 signature lincRNA, localized 139.5 kb upstream of the MAF gene [1].
Cellular Localization
linc-MAF-4, a chromatin associated CD4+ TH1 specific lincRNA [1].
Function
Linc-MAF-4 plays a key role in T lymphocyte differentiation [1].
Regulation
Linc-MAF-4 down-regulation skewed T cell differentiation toward TH2 [1].
A long-distance interaction between linc-MAF-4 and MAF genomic regions, where linc-MAF-4 associates with LSD1 and EZH2, suggesting linc-MAF-4 regulated MAF transcription by recruitment of chromatin modifiers [1].
Diseases
- allergy
- autoimmunity
- cancer
Expression
Sequencing data showed that high expression of linc-MAF-4 correlated with a low amount of MAF transcript in CD4+ TH1 cells, conversely TH2 cells had low expression of linc-MAF-4 and abundant MAF transcripts [1].
No correlation was observed between the expression linc-MAF-4 and its proximal upstream protein coding genes: CDYL2 and DYNLRB2 [1].
Sequence
>gi|559098521|ref|NR_104663.1| Homo sapiens MAF transcriptional regulator RNA (MAFTRR), long non-coding RNA
000081 AAGCGGGTGC CTCCTTCTAA TTCATCTTCA CCATTGTGGT TGTCAATATG GCATGCAGAA TTATCCCTCT TTCCAGGTTT 000160
000161 TCCCCAAGGA CCCTGGACAA TGCTGGTTTT TCCTATGTGC ACCACCAAAG GGCCTATCCA GCTTCTGGAG TCTACCTGGA 000240
000241 TTCCTGGGCT CATGGCCCCT TCCTCCATCT TCAAACCAGC AATGGAAATG GAAATGAAAG CAAAATGGAA GGACGTGAGG 000320
000321 TCCTTATCCT TGGAGATCCT ATGGAGTCTC CACAGACATT CCTTGAAGAA ATCAGATGGA GCTGAGCCTG ACACTCTAAC 000400
000401 CGGTTCTCTG GGTGTACTGC AAAGGTCTAA TAAAAGGCAC CTTTGCCCGG TTGCTTCAAT TCTAGGAAGC TGTTCAATGT 000480
000481 TTGACTATAT TTCTCCATAC TCATTTGAAA ACAGTTCATC AGGTTAGTTC AGTTGAATTA TTGCTCCTTG GGAGTTTTCC 000560
000561 AAACCCTGGA TTTCCTTCGG AGAGAGCTAG ATTCTATTCC ATTCTTGGAA TTCAGCTCCT TGCCCTTCTC TGTGACCCCG 000640
000641 GATCGCGAAT GCAGTAAAAA TAAAAATATC CTGCTGCCTC CTGCA
Labs working on this lncRNA
- Istituto Nazionale Genetica Molecolare “Romeo ed Enrica Invernizzi”, 20122 Milano, Italy [1].