Difference between revisions of "ENST00000414795.1"
| Line 19: | Line 19: | ||
===Regulation=== | ===Regulation=== | ||
LncRNA-CMPK2 is a bona fide ISG induced by both type I and type II IFNs<ref name="ref1" />. | LncRNA-CMPK2 is a bona fide ISG induced by both type I and type II IFNs<ref name="ref1" />. | ||
| − | |||
===Evolution=== | ===Evolution=== | ||
Revision as of 05:12, 30 June 2016
NRIR(also namedlncRNA-CMPK2), a spliced, polyadenylated nuclear transcript that was induced by IFN acts as a negative regulator of the IFN response.
Contents
Annotated Information
Name
NRIR: negative regulator of interferon response (non-protein coding)(HGNC nomenclature)
lncRNA-CMPK2[1]
Cellular Localization
- nuclear
Characteristics
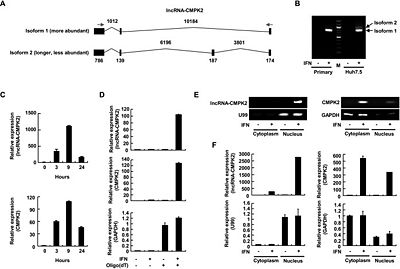
LncRNA-CMPK2, which was locacted on chromosome 2p25.2 and spans 832 bp long, was a spliced, polyadenylated nuclear transcript that was induced by IFN in diverse cell types from human and mouse[1].
Function
The IFN-induced lncRNA-CMPK2 acts as a negative regulator of the IFN response, at least partially through suppression of transcription of a subset of antiviral ISGs. Further, the level of lncRNA-CMPK2 was increased in liver samples from patients chronically infected with HCV[1].
Regulation
LncRNA-CMPK2 is a bona fide ISG induced by both type I and type II IFNs[1].
Evolution
IFN-mediated induction of lncRNA-CMPK2 is likely conserved among mammals and is not tissue-specific, but rather part of the global transcriptional response to IFN stimulation[1].
Labs working on this lncRNA
- Department of Biochemistry, School of Medicine, Case Western Reserve University, Cleveland, OH 44106, USA[1]
References
Basic Information
| Transcript ID |
ENST00000414795.1 |
| Source |
Gencode19 |
| Same with |
lnc-CMPK2-1:1,NONHSAT068918,NRIR,lncRNA-CMPK2 |
| Classification |
intergenic |
| Length |
506 nt |
| Genomic location |
chr2-:6969011..6980595 |
| Exon number |
4 |
| Exons |
6969011..6969086,6970099..6970237,6973445..6973561,6980422..6980595 |
| Genome context |
|
| Sequence |
000001 TTCAATCCCA CCCTTCCTTA CCAGATACTA AAGTCATTTC TACCTTTCCT CTTGAGATTT GCTGTTAGAT ATTAGAACTT 000080
000081 CTCTTTGTCA CATACTTACA GGGAGATTTT GATTATCCAT GGGATTCCAT TCATAACCTC ATAAACCACC CCCACGAAGA 000160 000161 AATTATATAT CTCGAAGAGG TTATATGGGC ACATTGTGAG ATGGTGGCCT CCCATGAGCC AAGAAAAGAG GGCTTAAAAT 000240 000241 GAATCCTACC TGGCTGGCAC CTTGATCTTG GACTTCCTAG CATCCAGAAA TGTGACATGG TTTTCTGGTG CCTTGGCAAC 000320 000321 TGCTCACGAT GCATGGGAAG ACTAAACACT TTGTGTACAG CATTCTGTCT CATCCAGTGA AGACTTCCTC TGGATTATTG 000400 000401 CAGCAGACAC CTCTAACCTC CTTAATCTGT ACTGAATCTT GCTCTGTCGC CAGGCTGGAG TGCAGTGGTG CGATATTGGC 000480 000481 TCACTGCAAC CTCCGCCTCT CGGGTT |
Predicted Small Protein
| Name | ENST00000414795.1_smProtein_296:379 |
| Length | 28 |
| Molecular weight | 3084.7697 |
| Aromaticity | 0.111111111111 |
| Instability index | 47.8222222222 |
| Isoelectric point | 8.41741943359 |
| Runs | 5 |
| Runs residual | 0.0214707461084 |
| Runs probability | 0.0334393216747 |
| Amino acid sequence | MVFWCLGNCSRCMGRLNTLCTAFCLIQ |
| Secondary structure | LEEEELLLLLHHHHHHHHHHHHHHHLL |
| PRMN | - |
| PiMo | - |