Difference between revisions of "MYCNOS"
| Line 3: | Line 3: | ||
MYCNOS | MYCNOS | ||
===Approved Name=== | ===Approved Name=== | ||
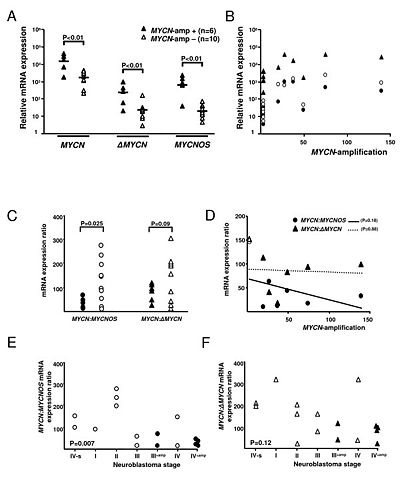
| − | [[File: Expression_level.jpg|right|thumb| | + | [[File: Expression_level.jpg|right|thumb|400px|'''Figure: MYCN, ΔMYCN, and MYCNOS mRNA expression levels in NBs.''' <ref name="ref1" />.]] |
MYCN opposite strand | MYCN opposite strand | ||
Revision as of 13:48, 30 June 2016
Contents
Annotated Information
Approved Symbol
MYCNOS
Approved Name
MYCN opposite strand
Synonyms
NCYM, N-CYM, MYCN-AS1
Chromosome
2p24.1
RefSeq ID
NR_026766
OMIM ID
605374
Ensembl ID
ENSG00000233718
pubmed IDs
1419902, 12880964, 19615087, 24391509
Characteristics
Both MYCNOS and ΔMYCN are potential inhibitors of MYCN either at the mRNA or at the protein level.[1].
Function
The MYCNOS:Neuroblastoma cells (NBs)carry an amplified MYCN gene are extremely malignant and MYCN-ratio in NBs is significantly correlated with both MYCN-amplification and NB-stage [1].
Regulation
MYCN and ΔMYCN are co-regulated [1].
Expression of the antisense gene MYCNOS might be relevant to the progression of NB, potentially by directly inhibiting MYCN transcription by transcriptional interference at the DNA level [1].
Diseases
- neuroblastoma
Expression
The ratio of MYCNOS:MYCN expression is directly correlated with NB disease stage (p = 0.007) [1].
Sequence
>gi|223029490|ref|NR_026766.1| Homo sapiens MYCN opposite strand (MYCNOS), transcript variant 2, long non-coding RNA
000081 CCTGGCTGCA GAATTCTAGC TCTCACGAGC ACGCAGACAA CCGCACTCGC AGCGGTGTGG GGCCGGCTGC TCAGGGGAAG 000160
000161 CCCCAGGCTC TCCGACCCAG CTACCGGGAA TGGGGCACCC TTTGGAGAAG AACCCCAGCC TGGGGTGGGG ACGCACCGGC 000240
000241 TCTCCGACAG CTCAAACACA GACAGATCTT CTAGAGCCGA GGGAATTTCT TTTCGCAGAA GCCATTACTC CCCCCGAGAG 000320
000321 AAGGCTGCAA AGCTGGGAAG CCCAGGGTGT GCTCCTCCCG CCCTTTTGGA CCCCCGGGCT TGCACCGGCT GCACTCTGAG 000400
000401 AACCAGCTGC GCGCGGAGCG GTGCAATGCA GCACCCACCC TGCGAGCCTG GCAATTGCTT GTCATTAAAA GAAAAAAAAA 000480
000481 TTACGGAGGG CTCCGGGGGT GTGTGTTGGG GAGGGGAGAC CGATGCTTCT AACCCAGCCC CCGCTTTGAC TGCGTGTTGT 000560
000561 GCAGCTGAGC GCGAGGCCAA CGTTGAGCAA GGCCTTGCAG GGAGGTTGCT CCTGTGTAAT TACGAAAGAA GGGTAGTCCG 000640
000641 AAGGTGCAAA ATAGCAGGGA GAGGACGCGC CCCCTTAGGA ACAAGACCTC TGGATGTTTC CAGTTTCAAA TTGAAAGAAG 000720
000721 AGGGGCGCCC CCCTTGTTTG AAAATAAATA AATAAATAAG TGCGAGCTAC
Labs working on this lncRNA
- Department of Human Genetics, Radboud University Nijmegen Medical Centre, Nijmegen Centre for Molecular Life Sciences, Donders Institute for Brain, Cognition and Behaviour, P.O. Box 9101, 6500 HB Nijmegen, the Netherlands.