Difference between revisions of "HTT-AS"
| Line 4: | Line 4: | ||
===Approved Name=== | ===Approved Name=== | ||
HTT antisense RNA (head to head) | HTT antisense RNA (head to head) | ||
| − | [[File: HTT-AS.jpg|right|thumb|400px|''' | + | [[File: HTT-AS.jpg|right|thumb|400px|'''HTT_AS1 expression.''' <ref name="ref1" />.]] |
===Previous Symbols=== | ===Previous Symbols=== | ||
HTT-AS1 | HTT-AS1 | ||
Revision as of 03:07, 26 September 2016
Contents
Annotated Information
Approved Symbol
HTT-AS
Approved Name
HTT antisense RNA (head to head)
Previous Symbols
HTT-AS1
Synonyms
HTTAS
Chromosome
4p16.3
RefSeq ID
NR_045414
Ensembl ID
ENSG00000251075
pubmed IDs
21672921, 23756188
Disease
- Huntington’s disease (HD)
Characteristics
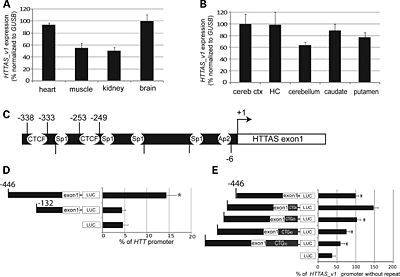
HTTAS is 5'capped, poly (A) tailed and contains three exons, alternatively spliced into HTT-AS1 (exons 1 and 3) and HTT-AS2 (exons 2 and 3)[1].
Expression
HTT-AS1 is expressed in multiple tissues types, with brain and heart expression 2-fold higher than muscle or kidney while is reduced in HD brains.[1].
Function
In cell systems, overexpression of HTT-AS1 specifically reduces endogenous HTT transcript levels, while siRNA knockdown of HTT-AS1 increases HTT transcript levels [1]. HTT-AS1 negatively regulates HTT expression [1]. Repeat length and HTT-AS1 interact to regulate HTT expression [1].
Labs working on this lncRNA
- Department of Psychiatry, Division of Neurobiology, Laboratory of Genetic Neurobiology [1].
References
Sequence
>gi|354623064|ref|NR_045414.1| Homo sapiens HTT antisense RNA (head to head) (HTT-AS), antisense RNA
000081 TGTAATGGGC AATGAACATG AACTATTCAG GGGAAGATTG TTAAATGAAA GAATCTCACT GGGCGGGGAG CTAAGCATGA 000160
000161 CACCTGGCCA CCTGCCTGTG CCTGTCCTGC CGAGAAGAGA GAAGAGGCCT TGAAAGCAGC CAGAGGGGGT GCACACCAGC 000240
000241 CTGGGCAGGG GGCAGTAAGC GGGAGCAGCC CCGACCCGAG ACCAGCAGCC CCTCTCTTCA CCTCCAAGCC GGAGCGTGGC 000320
000321 CTCATCCCTC CCAGGTGACT CTCTTCCCAG AAATGGTGCT GTCTCAAGAA GCCCCCTCTC TCCAAGCATC TGCCAGGCCG 000400
000401 TTTGAGCTGC CAGGAGTCTT TGTCCTGCCC CACCAGCTCT GTCTCATCTA GATGGGGCCT TCTGTCCAGT GCTACTGAAC 000480
000481 AGAATGTTCT GCATGCCTGG CCAGTGCAGA GACCTCCTTG CACCCCTGCT CCAAGGCTGG CTCTGCCACT GACAGATAGA 000560
000561 CTGACATTTG CACTGTTCTG AGCCCCTGGG GGTGCAGTAT GCAGAGCTAG GCCATCTCTG TCGTCCTACA AGAACCAGGC 000640
000641 AGCCCTGAGC AAGGCTTTTA ACATGCAAGG TGTACTCCAC TAGTCTTAAG ATATCTTGAG CCACTGTCTT GCTGCTCCCA 000720
000721 ACTCCCAAGC CACGCTGACT