Difference between revisions of "LINC-ROR"
Jiabao Cao (talk | contribs) (→Function) |
Jiabao Cao (talk | contribs) (→Function) |
||
| Line 20: | Line 20: | ||
RoR can also function as a negative regulator of p53 through interaction with an RNA binding protein, heterogeneous nuclear ribonucleoprotein I. Importantly, a 28-base RoR sequence carrying hnRNP I binding motifs is essential and suffcient for p53 repression. In addition, it can inhibit p53-mediated cell cycle arrest and apoptosis.<ref name="ref2" /> | RoR can also function as a negative regulator of p53 through interaction with an RNA binding protein, heterogeneous nuclear ribonucleoprotein I. Importantly, a 28-base RoR sequence carrying hnRNP I binding motifs is essential and suffcient for p53 repression. In addition, it can inhibit p53-mediated cell cycle arrest and apoptosis.<ref name="ref2" /> | ||
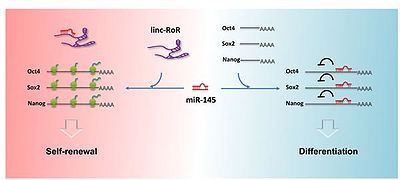
| − | Linc-RoR functions as an endogenous miRNA sponge for differentiation-related miRNAs. and linc-RoR, miRNAs, and the core TFs can form a regulatory circuit consisting of autoregulatory and dualnegative feedback loops during embryonic stem cell (ESC) self-renewal. This regulatory loop maintains a relative balance in self-renewing human embryonic stem cells (hESCs) to resist slight environmental changes and to elicit a rapid response to strong differentiation signals that promote hESCs differentiation. <ref name=" | + | Linc-RoR functions as an endogenous miRNA sponge for differentiation-related miRNAs. and linc-RoR, miRNAs, and the core TFs can form a regulatory circuit consisting of autoregulatory and dualnegative feedback loops during embryonic stem cell (ESC) self-renewal. This regulatory loop maintains a relative balance in self-renewing human embryonic stem cells (hESCs) to resist slight environmental changes and to elicit a rapid response to strong differentiation signals that promote hESCs differentiation. <ref name="ref3" /> |
| − | Linc-RoR modulates miR-145 levels, a sits overexpression diminishes endogenous miR-145 in self-renewing hESCs and drastically delays the increase in miR-145 upon hESC differentiation.<ref name=" | + | Linc-RoR modulates miR-145 levels, a sits overexpression diminishes endogenous miR-145 in self-renewing hESCs and drastically delays the increase in miR-145 upon hESC differentiation.<ref name="ref4" /> |
Linc-RoR can modulate cellular responses during hypoxic stress through modulation of HIF-1a and its downstream targets.<ref name="ref5" /> | Linc-RoR can modulate cellular responses during hypoxic stress through modulation of HIF-1a and its downstream targets.<ref name="ref5" /> | ||
Revision as of 13:43, 28 July 2017
Contents
Annotated Information
Name
LINC-ROR:long intergenic non-protein coding RNA, regulator of reprogramming[1]
Characteristics
The RoR gene is 2.6 kb in length, located in chromosome 18 (hg19 chr18:54,721,802-54,739,350), consisting of four exons. [2]
Cellular Localization
18q21.31
Function
LINC-ROR can modulate reprogramming of human differentiated cells to induce pluripotent stem cells.[1]
RoR can also function as a negative regulator of p53 through interaction with an RNA binding protein, heterogeneous nuclear ribonucleoprotein I. Importantly, a 28-base RoR sequence carrying hnRNP I binding motifs is essential and suffcient for p53 repression. In addition, it can inhibit p53-mediated cell cycle arrest and apoptosis.[2]
Linc-RoR functions as an endogenous miRNA sponge for differentiation-related miRNAs. and linc-RoR, miRNAs, and the core TFs can form a regulatory circuit consisting of autoregulatory and dualnegative feedback loops during embryonic stem cell (ESC) self-renewal. This regulatory loop maintains a relative balance in self-renewing human embryonic stem cells (hESCs) to resist slight environmental changes and to elicit a rapid response to strong differentiation signals that promote hESCs differentiation. [3]
Linc-RoR modulates miR-145 levels, a sits overexpression diminishes endogenous miR-145 in self-renewing hESCs and drastically delays the increase in miR-145 upon hESC differentiation.[4]
Linc-RoR can modulate cellular responses during hypoxic stress through modulation of HIF-1a and its downstream targets.[5]
Regulation
RoR-p53 autoregulatory feedback loop where p53 transcriptionally induces RoR expression.[2]
Linc-RoR transcription ias mainly controlled by the core TFs Oct4, Sox2, and Nanog.[4]
Diseases
malignant liver cancer [5]
Expression
RoR is highly expressed in embryonic stem cells and iPSCs.[2]
In self-renewing human embryonic stem cells (hESCs), linc-RoR is expressed at a high level and removes trace transcribed miRNAs when hESCs are subjected to temporary and slight differentiation agents.[3]
Linc-RoR expression is increased in hypoxic regions within tumor cell xenografts in vivo and is highly expressed in extracellular RNA released by hepatocellular cancer (HCC) cells during hypoxia[5]
Sequence
>NR_048536.1 Homo sapiens long intergenic non-protein coding RNA, regulator of reprogramming (LINC-ROR), long non-coding RNA
000081 TGACTCGGAT AGGGGGACCT CCCTTGGGAG ATCAGTACCC TGTCCTCCTG CTCTTTGCTC CGTGAGAAAG ATCCACCTAC 000160
000161 AACCTCAGGT CCTTAGACCA ACCAGCCCAA GAAACATCTC ACCAATTTCA AATCCAGACC CCACTGGAAA TCGGACTGTC 000240
000241 CAACTCACCT GACAGCCACT CCCACAGCCG CTGGAACTCT GGCCCAAGGC TCTCTGACTC CTTCCCAGAT CTTCTTGGCT 000320
000321 TAGCGGCTGA AGACTGACGC TGCCCGATCG CCTCGGAAGC CCCCTAGACC ATCACGGACG CCGAGCTTCG GGTAACTCTC 000400
000401 ACAGTGGAAG AAACACAACT ACAGATTTCT ACCTGGTGCA TGGCCATCGC TCATGAAGAC TACAACTTCC AGCTTCCTTT 000480
000481 GAAGAAAAAG AGGACTTGAT GGCATTGTCG CTAAGTAAGA AATAATGTGT GTGACTTCAG GGTTGCCCCC TTAAAGGGAG 000560
000561 GGGACATTTT CCATCCTGCT GTTCAGAGTA TGGATGTGAT GAGAGACATC TTGGATGATG CAGAGGAGGT GAACACCCCA 000640
000641 GGACAATGAA ACCACAGGAG AGAAAGAGCC TGCGCTGGCC CATCTAGCAC AGCCACTGGA CACAGGGACC ACCTGCCTCT 000720
000721 GCACTCTTAT GGAAGGAGGA AATCTAACTT TCCCAGTTTA AGGCACTGTT ACTTTGGGCT CCTGTTACAT AACTGTGGCA 000800
000801 GAATGAAGGT TCAACATGGA AACTGGCAAT GTTGAAGAAA CATAAAGTTC ATTGCTTGAA TATCTGAAGT TGTGGACTCA 000880
000881 ATCTCATACC TGCTCCACTT ATGAGTTATA GTTCTTCCAG GTCTCAGGAA TGGGATCAGC AGGTCTCAGG GTTGTACTCT 000960
000961 CCTGGATCTC TCACCAGCCA CCTCAAACCA GCTGCCATAG CCTGTCCACT TCCACTCCAA TCTTCTCTTC CTTCATCACC 001040
001041 TCCCTTGCAC ACCCTGATAA CCTCGAAAGA GAACTCTTCC CAAGGCTCTG TTCCAAACAC ATCGCCACTC TGCTTAGAAC 001120
001121 CTTCAATGAC TCCTCATGGC CTAGGAGGTT TCTCTCCCAT CTGGATCCAG CTGACGTTCC CAGCACCTTC TCCTGACTCC 001200
001201 TGTCTTTTCT TGAACCAGTT CTGCCCAACA AGGAGGAAAG GGCTGACAGA GTGAAAGTCC CAGGGCATGT GGGAATGTGA 001280
001281 CTCTTTTCAC TTTAAATTCT ATGACTGGAA AGTTTTGGGC AGAGTTGGAC ATGTGCACTT AGCTTCCAGA AGACAGAATC 001360
001361 CTTTTAAAAG AGTCAGAGAA AACACTGGCT TCCTGCCATG ACATGAGATA CAGACAGGAG AGTTGGGAAG CTTTTTAAAG 001440
001441 ATGGCACTAT GACTACAATC ACAGAAACTC TCCATGAGGA AGTAAAAGAA AGCACCTGCA ACACTCCAGC TATGCAGACC 001520
001521 ACTCTGTAAT GGGCTCAGAT CTGGACAGGT GTGTGGAAAG GTGGGTCAAC AGGTCAGGCG TCACAGACTT GGAACATTCA 001600
001601 TGGTGGAAAA GAAAAAGCCC CAAAGAAGAG ACTTCAGGAT AAATGAGAAA ATACTCAAGA CAGCAAAAGT CTCTTTTAGA 001680
001681 AATGTTGGAG AAAGAACACT TAATGTCAGG AGTTACTGTT GATTGATGGC CTTACTGTGT AGCAGGTGAG AAACCCATTG 001760
001761 TTCAGTTCCC TAAAGTCACC CTATTCTCCC AATCATCCTA TGGAGGGGGA ACCATGATGG TTATCCCCAT CTTATAAATA 001840
001841 AAGCAACAGA GGCTTAGAAG GACGAACTCT TTTTCTCAAG GTTACCCAGA TCATTTTGCA GAAGTCCCTA GATTTGAATC 001920
001921 ATGCTCTTGC TTTGAGGTTA AAGACACAGG GGAAGTCGAA CTCCTTATCT CCTATATCCT GAATAGGGAA AACCAAACAT 002000
002001 TGTCAAGAGG AGAGGAAGCC TGAGAGTTGG CATGAATCAG AGTGCTGGGC AGTCTGGAGT CTTCCCCACT GAGTTGATGA 002080
002081 TGGAACAGTA GAGTGGGGCC TGAGCCCCGT TAGGGCATGA GCTGCTGAAT GATTCATGTG AACACCATGC ACATGGGAGT 002160
002161 GAGGTTTTGA GCAGTGTGCC ACAGGAGCCT ACCCTCAGGC CCCACCATAA AATGTAGGGC CAGTCCTACA TTTTATCAAT 002240
002241 GACTTGCGTG AACACAGAAA ATGTGGATAC AACCAAAAGG TAACAATCCA ATTAAAAAAT GGACAAAAAA CTTGAATAAA 002320
002321 CATTTCTCAA AAGAAGATAT ACAAATGATC AAAAAGCATA TGAAAAAATG CTCAACATCG CTAATTGTAA GAGAAATGCA 002400
002401 AATCAAAACT ATAATGAGAT ACCACCTTAT ATTGATTAGG AAGACTGCTA TAAAAATAGT AAACAAACAA ACAAACAAAG 002480
002481 TAAGTCTTGG GGAGGATGCA GAGAAATTAA AATTTTTGTG CACTGTTAGT GGGAATGTAA AATGGTGCAG CTGTTACGGG 002560
002561 AAACGGTATG ACAGTTCCTC AAAAAATAAA A
Labs working on this lncRNA
- Stem Cell Transplantation Program, Division of Pediatric Hematology and Oncology, Manton Center for Orphan Disease Research, Children's Hospital Boston and Dana Farber Cancer Institute, Boston, Massachusetts, USA.[1]
- Department of Medical Microbiology, Immunology and Cell Biology, Southern Illinois University School of Medicine, Springfield, IL 62794-9621, USA.[2]
- Department of Histology and Embryology, College of Basic Medicine, Second Military Medical University, Shanghai 200433, China.[3]
- Yale Stem Cell Center and Department of Cell Biology, Yale University School of Medicine, New Haven, CT 06509, USA.[4]
- Mayo Clinic, Jacksonville, FL 32224, USA.[5]
References
- ↑ 1.0 1.1 1.2 Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010 Dec;42(12):1113-7.
- ↑ 2.0 2.1 2.2 2.3 2.4 Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, Lu Z, Bai C, Watabe K, Mo YY. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013 Mar;23(3):340-50.
- ↑ 3.0 3.1 3.2 3.3 Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013 Apr 15;25(1):69-80.
- ↑ 4.0 4.1 4.2 Cheng EC, Lin H. Repressing the repressor: a lincRNA as a MicroRNA sponge in embryonic stem cell self-renewal. Dev Cell. 2013 Apr 15;25(1):1-2. doi: 10.1016/j.devcel.2013.03.020.
- ↑ 5.0 5.1 5.2 5.3 Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci. 2014 Apr 1;127(Pt 7):1585-94. doi: 10.1242/jcs.141069. Epub 2014 Jan 24.