Difference between revisions of "XACT"
| (One intermediate revision by the same user not shown) | |||
| Line 22: | Line 22: | ||
===Evolution=== | ===Evolution=== | ||
| − | Although the organization of the genomic region encompassing AMOT and HTR2C is well conserved in placental mammals and marsupials, the sequence between these two genes shows moderate conservation, with several conserved blocks present in placental mammals but not marsupials. The LTR that corresponds to the 5' end of XACT is conserved in chimpanzees but not macaques or more distally related species, suggesting that the insertion of these LTR elements is a very recent event <ref name="ref1" />. | + | Although the organization of the genomic region encompassing AMOT and HTR2C is well conserved in placental mammals and marsupials, the sequence between these two genes shows moderate conservation, with several conserved blocks present in placental mammals but not marsupials. The LTR (long terminal repeat) that corresponds to the 5' end of XACT is conserved in chimpanzees but not macaques or more distally related species, suggesting that the insertion of these LTR elements is a very recent event <ref name="ref1" />. |
| − | No XACT-like transcript expression is detected in mouse, which indicates that XACT might not be conserved in | + | No XACT-like transcript expression is detected in mouse, which indicates that XACT might not be conserved in mouse <ref name="ref1" />. |
===Expression=== | ===Expression=== | ||
| Line 34: | Line 34: | ||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
| − | * University Paris Diderot, Sorbonne Paris City, Epigenetics and Cell Fate, Paris, France | + | * University Paris Diderot, Sorbonne Paris City, Epigenetics and Cell Fate, Paris, France |
==References== | ==References== | ||
<references> | <references> | ||
| − | <ref name="ref1">Vallot, C., | + | <ref name="ref1">Vallot, C., Huret, C., Lesecque, Y., Resch, A., Oudrhiri, N., Bennaceur-Griscelli, A., Duret, L. and Rougeulle, C. (2013) XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nat Genet, 45, 239-241.</ref>(1) |
| − | </ref>(1) | ||
</references> | </references> | ||
Latest revision as of 08:22, 26 September 2017
Contents
Annotated Information
Approved Symbol
XACT: X active specific transcript
Chromosome
Xq23
RefSeq ID
NR_131204
OMIM ID
300901
Ensembl ID
ENSG00000241743
Characteristics
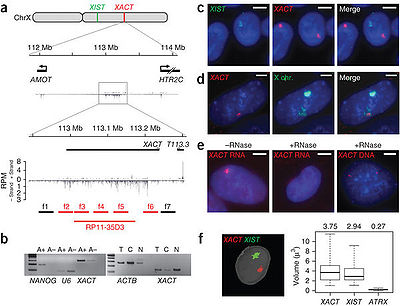
XACT is a 251.8-kb transcription unit(112,983,323–113,235,148 bp) and it is located on chromosome Xq23 between the protein-coding genes AMOT and HTR2C [1]. XACT is polyadenylated and belongs to intergenic lncRNA group [1].
Cellular Location
XACT is mostly nuclear [1].
Function
XACT is involved in the control of XCI (X-chromosome inactivation) initiation in human [1]. XACT is expressed from and coats the active X chromosome specifically in human pluripotent cells. In the absence of XIST, XACT is expressed from both X chromosomes in human but not in mouse, suggesting a unique role for XACT in the control of human XCI initiation [1].
Evolution
Although the organization of the genomic region encompassing AMOT and HTR2C is well conserved in placental mammals and marsupials, the sequence between these two genes shows moderate conservation, with several conserved blocks present in placental mammals but not marsupials. The LTR (long terminal repeat) that corresponds to the 5' end of XACT is conserved in chimpanzees but not macaques or more distally related species, suggesting that the insertion of these LTR elements is a very recent event [1].
No XACT-like transcript expression is detected in mouse, which indicates that XACT might not be conserved in mouse [1].
Expression
XACT is expressed in active X chromosome in human but not in mouse, and the expression of XACT is restricted to pluripotent and early differentiating cells in human [1].
Sequence
>gi|784642207|ref|NR_131204.1| Homo sapiens X active specific transcript (non-protein coding) (XACT), long non-coding RNA
000081 TGCTGATGAT GTCCTAAACC TGATCACTTG TCTCAAAATT TAGAAAGTTA ATGGCTGAAG ATTCAGCAGC CATTCTGAAC 000160
000161 TGGAAATTAC ATGTTAAGGC TGTTATAGAA GAAATAAAGA AATTAAATCT CTATGTCCCC AAGGTATCTC TATACCGGCC 000240
000241 TGGAATGCCT ATCTCCAAAC CTCCTTTACA TGAGACAGAA ACAGCTTCTA TCACCTTAAA ATCTGTTACT TTAGGTTTTT 000320
000321 TTCATTATGA GCAGCTAAAA TGAATCTTAC TTGATACAGT GTGAGTGTTC ATGGATTAGG GCATTGTTTG ATAAGAGGAG 000400
000401 TAAGAAAAGA ATATAAATTG TATTGGCCCT GATATGCCAC AAACAATGAG GGCTGAAGCC AAATTAATCA AAGAATAAAC 000480
000481 TCAAATACAA AGAACAAAAA AAAAAAAAA
Labs working on this lncRNA
- University Paris Diderot, Sorbonne Paris City, Epigenetics and Cell Fate, Paris, France