Difference between revisions of "MDS2"
(Created page with "==Annotated Information== ===Approved Symbol=== MDS2 ===Approved Name=== myelodysplastic syndrome 2 translocation associated ===Previous Symbols=== _ ===Synonyms=== _ ===Chrom...") |
|||
| Line 1: | Line 1: | ||
==Annotated Information== | ==Annotated Information== | ||
===Approved Symbol=== | ===Approved Symbol=== | ||
| − | MDS2 | + | MDS2 (myelodysplastic syndrome 2 translocation associated) |
| − | |||
| − | myelodysplastic syndrome 2 translocation associated | ||
| − | |||
| − | |||
| − | |||
| − | |||
===Chromosome=== | ===Chromosome=== | ||
1p36 | 1p36 | ||
| − | |||
| − | |||
===OMIM ID=== | ===OMIM ID=== | ||
607305 | 607305 | ||
| Line 18: | Line 10: | ||
===pubmed IDs=== | ===pubmed IDs=== | ||
12203785 | 12203785 | ||
| + | [[File:Analysis of the expression levels of genes surrounding MDS2 in the patient and in three unrelated normal bone marrow samples.png|right|thumb|400px|''' Analysis of the expression levels of genes surrounding MDS2 in the patient and in three unrelated normal bone marrow samples'''<ref name="ref1" />]] | ||
| + | |||
| + | ===Characteristics=== | ||
| + | The MDS2 gene is approximately 13 kb and is flanked by E2F2 and ID3 at its 5� end (telomeric) and by RPL11 and TCEB3 at its 3�' end (centromeric) and gives rise to four alternatively spliced products.<ref name="ref1" /> | ||
| + | The predicted ETV6-MDS2 fusion product contains 58 amino acids, starting at the first ATG of ETV6 and ending in aTGA stop codon only four amino acids after ETV6 exon 2.<ref name="ref1" /> | ||
| + | |||
| + | ===Expression=== | ||
| + | Oligonucleotide primers' sequences of MDS2 are showed as follow. | ||
| + | {| class='wikitable' style="text-align:center" | ||
| + | |- | ||
| + | ! | Primer | ||
| + | ! | Sequences | ||
| + | |- | ||
| + | | | NG-R1 | ||
| + | | | GCGACAGCCAGGTCTTAA <ref name="ref1" /> | ||
| + | |- | ||
| + | | | NGex1-F1 | ||
| + | | | CCGCCTGTATCCGTGCCTGTT <ref name="ref1" /> | ||
| + | |- | ||
| + | | | NGex6-F1 | ||
| + | | | AGCAGTCCAGCTATGAGGGGAGGT <ref name="ref1" /> | ||
| + | |- | ||
| + | | | NGex6-R1 | ||
| + | | | TGGAGGCGACAGCCAGGTCTTAA <ref name="ref1" /> | ||
| + | |- | ||
| + | | | NGex6-F2 | ||
| + | | | GACCCTGGAGAGGCCATTAAGACCT <ref name="ref1" /> | ||
| + | |- | ||
| + | | | NGex6-R2 | ||
| + | | | GTCTTAATGGCCTCTCCAGGGTCAG <ref name="ref1" /> | ||
| + | |} | ||
| + | |||
| + | ===Disease=== | ||
| + | MyelodysplasticSyndrome <ref name="ref1" /> | ||
| + | |||
| + | ==Labs working on this lncRNA== | ||
| + | * Department of Genetics, University of Navarra, Pamplona, Spain. | ||
| + | |||
| + | ==References== | ||
| + | <references> | ||
| + | <ref name="ref1"> Odero, M.D., et al., A novel gene, MDS2, is fused to ETV6/TEL in a t(1;12)(p36.1;p13) in a patient with myelodysplastic syndrome. Genes Chromosomes Cancer, 2002. 35(1): p. 11-9. | ||
| + | </ref>(1) | ||
| + | </references> | ||
Revision as of 13:54, 13 November 2017
Contents
Annotated Information
Approved Symbol
MDS2 (myelodysplastic syndrome 2 translocation associated)
Chromosome
1p36
OMIM ID
607305
RefSeq(supplied by NCBI)
NR_027042
pubmed IDs
12203785
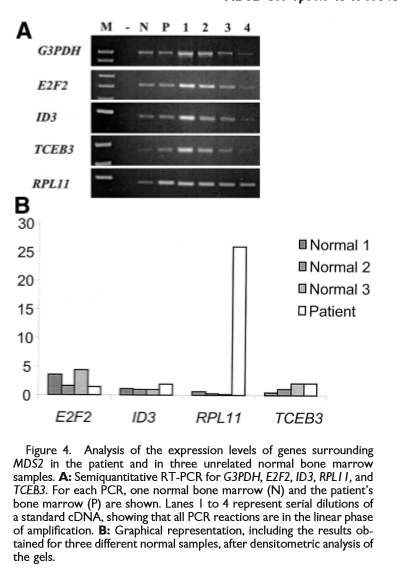
Analysis of the expression levels of genes surrounding MDS2 in the patient and in three unrelated normal bone marrow samples[1]
Characteristics
The MDS2 gene is approximately 13 kb and is flanked by E2F2 and ID3 at its 5� end (telomeric) and by RPL11 and TCEB3 at its 3�' end (centromeric) and gives rise to four alternatively spliced products.[1] The predicted ETV6-MDS2 fusion product contains 58 amino acids, starting at the first ATG of ETV6 and ending in aTGA stop codon only four amino acids after ETV6 exon 2.[1]
Expression
Oligonucleotide primers' sequences of MDS2 are showed as follow.
| Primer | Sequences |
|---|---|
| NG-R1 | GCGACAGCCAGGTCTTAA [1] |
| NGex1-F1 | CCGCCTGTATCCGTGCCTGTT [1] |
| NGex6-F1 | AGCAGTCCAGCTATGAGGGGAGGT [1] |
| NGex6-R1 | TGGAGGCGACAGCCAGGTCTTAA [1] |
| NGex6-F2 | GACCCTGGAGAGGCCATTAAGACCT [1] |
| NGex6-R2 | GTCTTAATGGCCTCTCCAGGGTCAG [1] |
Disease
MyelodysplasticSyndrome [1]
Labs working on this lncRNA
- Department of Genetics, University of Navarra, Pamplona, Spain.