Difference between revisions of "DANT1"
Chunlei Yu (talk | contribs) (Created page with "==Annotated Information== ===Approved Symbol=== DANT1 ===Approved Name=== DXZ4 associated non-coding transcript 1, proximal ===Previous Symbols=== ===Synonyms=== _ ===Chromos...") |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Annotated Information== | ==Annotated Information== | ||
===Approved Symbol=== | ===Approved Symbol=== | ||
| − | DANT1 | + | DANT1 (DXZ4 associated non-coding transcript 1, proximal) |
| − | + | [[File: ''Characterization of the DANT1 and DANT2 lncRNAs.jpg|right|thumb|400px|'''Characterization of the DANT1 and DANT2 lncRNAs''' <ref name="ref1" />.]] | |
| − | DXZ4 associated non-coding transcript 1, proximal | ||
| − | = | ||
| − | |||
| − | |||
| − | |||
===Chromosome=== | ===Chromosome=== | ||
Xq23 | Xq23 | ||
| − | |||
| − | |||
===Ensembl ID=== | ===Ensembl ID=== | ||
ENSG00000229335 | ENSG00000229335 | ||
| − | === | + | ===Characteristics=== |
| − | + | DANT1 generates an array-traversing transcript (ATT) with the longest DANT1 transcript entirely contained within, but anti-sense to, the longest predicted DANT2 transcript.<ref name="ref1" /> | |
| + | DANT1 produces both non-ATT and ATT transcript isoforms.<ref name="ref1" /> | ||
| + | Low levels of H3K9me3 can be observed at the DANT1 promoter in hESC.<ref name="ref1" /> | ||
| + | |||
| + | ===Function=== | ||
| + | DANT1 lncRNA is linked to changes in DXZ4 chromatin.<ref name="ref1" /> | ||
| + | |||
| + | ===Expression=== | ||
| + | The short DANT1 transcript detected in hESCs was of particularly low abundance, and could only be confidently detected in prostate, testis, trachea and spinal cord.<ref name="ref1" /> | ||
| + | |||
| + | ==Labs working on this lncRNA== | ||
| + | * Department of Biological Science, Florida State University, 319 Stadium Drive, King 3076, Tallahassee, FL, 32306-4295, USA.<ref name="ref1" /> | ||
| + | * NHLBI, National Institutes of Health, 10 center Drive, Building 10 Rm 6D12, Bethesda, MD, 20892, USA.<ref name="ref1" /> | ||
| + | * Department of Biological Science, Florida State University, 319 Stadium Drive, King 3076, Tallahassee, FL, 32306-4295, USA. <ref name="ref1" /> | ||
| + | |||
| + | ==References== | ||
| + | <references> | ||
| + | <ref name="ref1"> 1: Figueroa DM, Darrow EM, Chadwick BP. Two novel DXZ4-associated long noncoding | ||
| + | RNAs show developmental changes in expression coincident with heterochromatin | ||
| + | formation at the human (Homo sapiens) macrosatellite repeat. Chromosome Res. 2015 | ||
| + | Dec;23(4):733-52. doi: 10.1007/s10577-015-9479-3. | ||
| + | </ref>(1) | ||
| + | </references> | ||
Latest revision as of 13:55, 2 December 2017
Contents
Annotated Information
Approved Symbol
DANT1 (DXZ4 associated non-coding transcript 1, proximal)
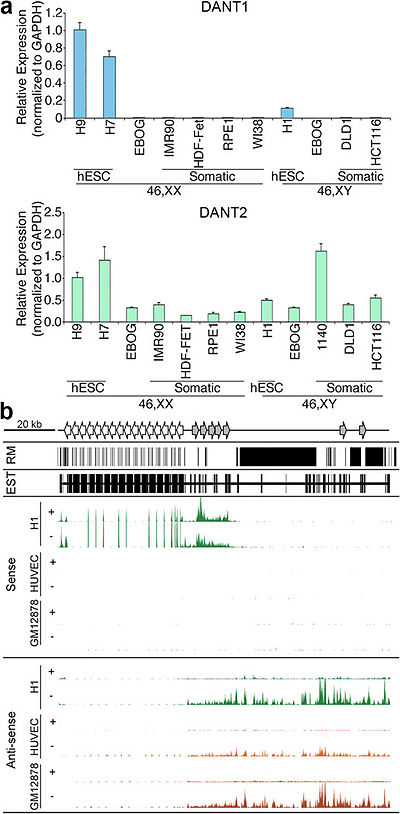
Characterization of the DANT1 and DANT2 lncRNAs [1].
Chromosome
Xq23
Ensembl ID
ENSG00000229335
Characteristics
DANT1 generates an array-traversing transcript (ATT) with the longest DANT1 transcript entirely contained within, but anti-sense to, the longest predicted DANT2 transcript.[1] DANT1 produces both non-ATT and ATT transcript isoforms.[1] Low levels of H3K9me3 can be observed at the DANT1 promoter in hESC.[1]
Function
DANT1 lncRNA is linked to changes in DXZ4 chromatin.[1]
Expression
The short DANT1 transcript detected in hESCs was of particularly low abundance, and could only be confidently detected in prostate, testis, trachea and spinal cord.[1]
Labs working on this lncRNA
- Department of Biological Science, Florida State University, 319 Stadium Drive, King 3076, Tallahassee, FL, 32306-4295, USA.[1]
- NHLBI, National Institutes of Health, 10 center Drive, Building 10 Rm 6D12, Bethesda, MD, 20892, USA.[1]
- Department of Biological Science, Florida State University, 319 Stadium Drive, King 3076, Tallahassee, FL, 32306-4295, USA. [1]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1: Figueroa DM, Darrow EM, Chadwick BP. Two novel DXZ4-associated long noncoding RNAs show developmental changes in expression coincident with heterochromatin formation at the human (Homo sapiens) macrosatellite repeat. Chromosome Res. 2015 Dec;23(4):733-52. doi: 10.1007/s10577-015-9479-3.