Difference between revisions of "ENST00000442621.1"
| (One intermediate revision by the same user not shown) | |||
| Line 13: | Line 13: | ||
===Cellular Localization=== | ===Cellular Localization=== | ||
Its subcellular localization was predominantly cytoplasmic<ref name="ref1" />. | Its subcellular localization was predominantly cytoplasmic<ref name="ref1" />. | ||
| + | |||
| + | ===Disease=== | ||
| + | ovarian cancer<ref name="ref1" />. | ||
===Function=== | ===Function=== | ||
Latest revision as of 06:13, 3 April 2018
OVAAL, 1489 nt ovarian adenocarcinoma amplified long non-coding RNA, which is located at the center of a narrowly amplified intergenic segment, contains three annotated exons, appears to have a cytoplasmic non-coding function.
Contents
Annotated Information
Name
OVAAL: ovarian adenocarcinoma amplified long non-coding RNA(HGNC nomenclature)
LINC01131, "long intergenic non-protein coding RNA 1131", OVAL, "ovarian adenocarcinoma amplified lncRNA"[1]
Characteristics
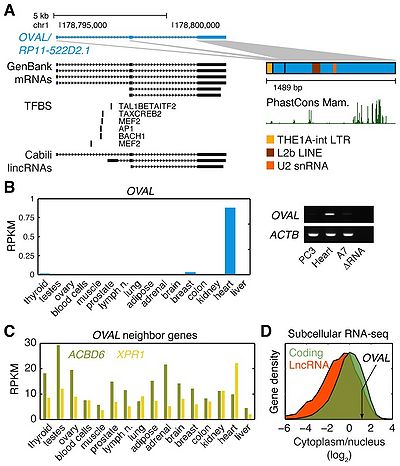
The OVAL gene, which is located at the center of a narrowly amplified intergenic segment that lacks other annotated genes, contains three annotated exons that give rise to a predicted 1489 nt non-coding RNA, where the large third exon contributes most of the sequence. Many of theOVAL ESTs and mRNAs originated from human melanoma cells[1].
Cellular Localization
Its subcellular localization was predominantly cytoplasmic[1].
Disease
ovarian cancer[1].
Function
OVAL appears to have a cytoplasmic non-coding function that is independent of its protein-coding neighbors[1].
Expression
Reverse transcription PCR
| Primer | Forward | Reverse |
|---|---|---|
| OVAL | 5'-AGGCCAATATGCAGACAAGG-3' | 5'-AGTTCTCCAGTGGGGGTCTT-3'[1] |
| ACTB | 5'-ACTCTTCCAGCCTTCCTTCC-3' | 5'-GTACTTGCGCTCAGGAGGAG-3'[1] |
Evolution
Evolutionary conservation,specific regions in the last exon demonstrated elevated conservation.
Labs working on this lncRNA
- Department of Medical Biochemistry & Cell Biology, Institute of Biomedicine, The Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden[1]
References
Basic Information
| Transcript ID |
ENST00000442621.1 |
| Source |
Gencode19 |
| Same with |
lnc-XPR1-2:2, OVAAL, LINC01131, OVAL |
| Classification |
intergenic |
| Length |
1489 nt |
| Genomic location |
chr1+:180528112..180535654 |
| Exon number |
3 |
| Exons |
180528112..180528191,180531340..180531480,180534387..180535654 |
| Genome context |
|
| Sequence |
000001 TGCCACTGCC ATGTAAGAAG TGCCTTTCAC CACCCCCACA CCCTGCCATG ACTCTGAGGC CTCCCCAGCC ACGTGGAACT 000080
000081 GAGGCAGCTG TATGGAGGCC AATATGCAGA CAAGGGAAGT AACTCAGGCA TTTTGGGGGA GAGGTACAGA AAGAAGAGAA 000160 000161 AAGACTGAAC TGACTGCTGT TCTGAACAAG AACTGTGACT ATATGTGGTA CTCCCAGAAT GCTGGTTTCC TTTCAGACCC 000240 000241 ATCAAGTGGC CATCCTATTG GCTTGGACTG AAGACCCCCA CTGGAGAACT GTCTCTGCAG CCTATACTGA TGACACTGGC 000320 000321 CACTGTCCAG GGCTCCACCC CAAGGAGTCC AAGCCCCAGA CATTTAGCCT TAGAGAGAAT CCAGATTTTG CATTTCCAGT 000400 000401 GCAGGGATTG ATGGAGCCTT GTGTATCAAA GTGCCAATTT CTTTCTAACA AGCTAGGGAG GATACCGTTG ACAGCAGCAG 000480 000481 AAAGGGGTAG CAGTGGTTTG CTTTCCAAAT GGTAATAAGA AGGGCCTGGC AGTGGCAACC TGTCCACGTA TCATTAAAAA 000560 000561 TGGACTCTCC TGCTTCGTTT GGAATTCAAC CCAATGAACT TGAATAAACT CTGGACAATG ACCAGCACAG AGCCCTAAGC 000640 000641 AGGATACAGC AAGATTATTT GCTTTACTCA TCCACTGAGT ACCTACCATG TGCCAAATAC TATGCTATAT GCCGAGTATA 000720 000721 CACTGGCAAA TAAAACAGAT ATGGTCCCTG GAGCCTATGA ATCTTACCCC ATCTAGCCAC AAATAATAAA AATAAGAAGA 000800 000801 ATCTGAAGGA CAAGTGGGAC AGAAATACTC AAGCAGAATG AGGGTGAGAT TGAAGTCCCT GAACATACAG TGCCCTTTAC 000880 000881 ACTTGATCTT AGCCAAAAGG CCGAGAAGAG ATTACAGTGC TTTTTAAAGG TAGCATTTCA CCCAGCCAAT AATCAATCCC 000960 000961 TTTGGCCTTC TTGAGGTGCA AGGCCACACA CTCAAATTCT TTAATCAAAC AATAGGCTAG TGCAAATGGA GCTATTTCTT 001040 001041 AAACCCAGCT GTTTACACAG CAGCTACAAA TCTGTTCAGA AGGAATCCCC CACCATCCAC AACTGTCTAT CGAGCAGATG 001120 001121 TGCAAGAGTA TTTGTCTCTC ACCACCAGGG TTGCAGTCAG GACAAGAGAG CAGGTTTGAA CGTCTTCGGC TCCTAGAACT 001200 001201 GCACAAGCTG TGTCTTCAGT GCCCAGAGGC GGCTGCAACG TCTCCCCAGT TCTCCTCTCA GAACAATGGA AATTTTTTTT 001280 001281 AGAGGCGTAA TAGGAAGGTT TGCATTGAAT TATTAAACAA TGGGTATTGA AAAATATGTT TTCTTTGACC TCTGTACTTC 001360 001361 TAGCTTTTAC CAGTCAGTGT TTTCTTGTAC CAAGGCTAAG ATATGTATAA CACTGAAATG GATGTCTTCT TATATAATAT 001440 001441 TTTACAAGCT TTTGAAAACC TGTGCCCTTT AATAAATTTT AAAACGTTT |