Difference between revisions of "CCAT2"
Chunlei Yu (talk | contribs) |
|||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | ''CCAT2'', a novel long noncoding RNA transcript (lncRNA) harboring a single nucleotide polymorphism rs6983267 SNP, | + | ''CCAT2'', a novel long noncoding RNA transcript (lncRNA) harboring a single nucleotide polymorphism rs6983267 SNP, promoting tumor growth, metastasis, and chromosomal instability. <ref name="ref1" /> <ref name="ref2" /> |
==Annotated Information== | ==Annotated Information== | ||
===Name=== | ===Name=== | ||
''CCAT2'', colon cancer-associated transcript 2 (CCAT2) (HGNC nomenclature) | ''CCAT2'', colon cancer-associated transcript 2 (CCAT2) (HGNC nomenclature) | ||
| + | LINC00873, "long intergenic non-protein coding RNA 873", NCCP1, "non-coding RNA involved in cancer predisposition 1"<ref name="ref1" />. | ||
===Characteristics=== | ===Characteristics=== | ||
| − | ''CCAT2'' is transcribed from chromosome 8q24 | + | ''CCAT2'' is transcribed from chromosome 8q24 which is a very conserved region harboring a single nucleotide polymorphism rs6983267 <ref name="ref2" />. |
===Function=== | ===Function=== | ||
[[File: A model of CCAT2 locus involvement in CRC.jpg|right|thumb|400px|'''A model of ''CCAT2'' locus involvement in CRC''' <ref name="ref1" />.]] | [[File: A model of CCAT2 locus involvement in CRC.jpg|right|thumb|400px|'''A model of ''CCAT2'' locus involvement in CRC''' <ref name="ref1" />.]] | ||
| − | ''CCAT2'' promotes cancer growth and metastasis and induces chromosomal instability | + | ''CCAT2'' which responds to WNT signaling can regulates MYC transcription and promotes cancer growth and metastasis and induces chromosomal instability <ref name="ref1" />. |
| − | ''CCAT2'' is a lung adenocarcinoma-specific lncRNA and promotes invasion of NSCLC and highlight its potential as a biomarker for lymph node metastasis | + | |
| + | ''CCAT2'' is a lung adenocarcinoma-specific lncRNA and promotes invasion of NSCLC and highlight its potential as a biomarker for lymph node metastasis <ref name="ref2" />. | ||
| + | |||
===Regulation=== | ===Regulation=== | ||
| − | rs6983267 status affects the ''CCAT2'' expression, providing an additional mechanism linking the risk allele of rs6983267 with higher CRC risk | + | rs6983267 status affects the ''CCAT2'' expression, providing an additional mechanism linking the risk allele of rs6983267 with higher CRC risk. <ref name="ref1" /> |
===Diseases=== | ===Diseases=== | ||
| − | * | + | *Breast cancer <ref name="ref3" /> |
| + | *Colon cancer<ref name="ref1" /> | ||
| + | *Gastric cancer <ref name= "ref4" /> | ||
| + | *Non-small cell lung cancer<ref name="ref2" /> | ||
===Expression=== | ===Expression=== | ||
| Line 64: | Line 70: | ||
| | GATCCCCTTAACCTCTTCCTATCTCATTCAAGAGATGAGATAGGAAGAGGTTAATTTTTA<ref name="ref1" /> | | | GATCCCCTTAACCTCTTCCTATCTCATTCAAGAGATGAGATAGGAAGAGGTTAATTTTTA<ref name="ref1" /> | ||
|} | |} | ||
| + | |||
| + | ==Labs working on this lncRNA== | ||
| + | * Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA; <ref name="ref1" /> | ||
| + | * Jiangsu Key Laboratory of Molecular and Translational Cancer Research, Nanjing Medical University Affiliated Cancer Hospital, Cancer Institute of Jiangsu Province, Nanjing, 210009, China <ref name="ref2" />. | ||
| + | * Department of Pathology, Josephine Nefkens Institute, Erasmus Medical Center, Rotterdam, The Netherlands | ||
| + | |||
| + | ==References== | ||
| + | <references> | ||
| + | <ref name="ref1"> Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer[J]. Genome research. 2013,23(9):1446-61. | ||
| + | </ref>(1) | ||
| + | <ref name="ref2"> Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer[J]. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014,35(6):5375-80. | ||
| + | </ref>(2) | ||
| + | <ref name="ref3"> Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R et al. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations[J]. Oncotarget. 2013, 4(10):1748. | ||
| + | </ref>(3) | ||
| + | <ref name="ref4"> Wang C-Y, Hua L, Yao K-H, Chen J-T, Zhang J-J & Hu J-H. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis[J]. International journal of clinical and experimental pathology. 2015, 8(1):779. | ||
| + | </ref>(4) | ||
| + | </references> | ||
| + | |||
===Sequence=== | ===Sequence=== | ||
| Line 93: | Line 117: | ||
TATCCTGCAGGTGTTCAGCTTCCCCTGTTCTACTCTGGGAAGAGAGCCGTGGGCAACATCAGCCCAGAAG | TATCCTGCAGGTGTTCAGCTTCCCCTGTTCTACTCTGGGAAGAGAGCCGTGGGCAACATCAGCCCAGAAG | ||
AC</dnaseq> | AC</dnaseq> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 06:42, 14 November 2018
CCAT2, a novel long noncoding RNA transcript (lncRNA) harboring a single nucleotide polymorphism rs6983267 SNP, promoting tumor growth, metastasis, and chromosomal instability. [1] [2]
Contents
Annotated Information
Name
CCAT2, colon cancer-associated transcript 2 (CCAT2) (HGNC nomenclature) LINC00873, "long intergenic non-protein coding RNA 873", NCCP1, "non-coding RNA involved in cancer predisposition 1"[1].
Characteristics
CCAT2 is transcribed from chromosome 8q24 which is a very conserved region harboring a single nucleotide polymorphism rs6983267 [2].
Function
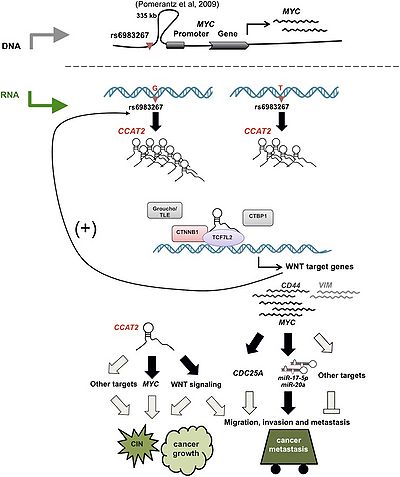
CCAT2 which responds to WNT signaling can regulates MYC transcription and promotes cancer growth and metastasis and induces chromosomal instability [1].
CCAT2 is a lung adenocarcinoma-specific lncRNA and promotes invasion of NSCLC and highlight its potential as a biomarker for lymph node metastasis [2].
Regulation
rs6983267 status affects the CCAT2 expression, providing an additional mechanism linking the risk allele of rs6983267 with higher CRC risk. [1]
Diseases
Expression
CCAT2 is expressed at high levels in MSS CRC tumor tissues and influences MYC levels, but has very low-level expression in normal colon tissues. [1]
| PCR Primers | Sequence |
|---|---|
| CCAT2 F1 | CCCTGGTCAAATTGCTTAACCT[1] |
| CCAT2 R1 | TTATTCGTCCCTCTGTTTTATGGAT[1] |
| CCAT2 F2 | CCGAGGTGATCAGGTGGACTTTC[1] |
| CCAT2 R2 | GTCTTCTGGGCTGATGTTGC[1] |
| siRNA | Target Sequence |
| CCAT2 siRNA 1 | TTAACCTCTTCCTATCTCA[1] |
| CCAT2 siRNA 2 | AGGTGTAGCCAGAGTTAAT[1] |
| CCAT2 siRNA 3 | CGAATAAACTCTCCTCCTA[1] |
| shRNA | Sequences cloned into pRS vector |
| CCAT2 shRNA1 | GATCCCCAATTGCTTAACCTCTTCCTTTCAAGAGAAGGAAGAGGTTAAGCAATTTTTTTA[1] |
| CCAT2 shRNA2 | GATCCCCGATGAAAGGCACTGAGAAATTCAAGAGATTTCTCAGTGCCTTTCATCTTTTTA[1] |
| CCAT2 shRNA3 | GATCCCCTTAACCTCTTCCTATCTCATTCAAGAGATGAGATAGGAAGAGGTTAATTTTTA[1] |
Labs working on this lncRNA
- Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA; [1]
- Jiangsu Key Laboratory of Molecular and Translational Cancer Research, Nanjing Medical University Affiliated Cancer Hospital, Cancer Institute of Jiangsu Province, Nanjing, 210009, China [2].
- Department of Pathology, Josephine Nefkens Institute, Erasmus Medical Center, Rotterdam, The Netherlands
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer[J]. Genome research. 2013,23(9):1446-61.
- ↑ 2.0 2.1 2.2 2.3 2.4 Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer[J]. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014,35(6):5375-80.
- ↑ Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R et al. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations[J]. Oncotarget. 2013, 4(10):1748.
- ↑ Wang C-Y, Hua L, Yao K-H, Chen J-T, Zhang J-J & Hu J-H. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis[J]. International journal of clinical and experimental pathology. 2015, 8(1):779.
Sequence
>gi|565671809|ref|NR_109834.1| Homo sapiens colon cancer associated transcript 2 (non-protein coding) (CCAT2), long non-coding RNA
000081 AGAAAGCTAG TATGAGACTA CCAAATGATC ATCAGACATT TCTTGAACAC CAATTAAATT GCTAGGTATG CTAAAGTTTG 000160
000161 CAAAACTGGT ATAGACACCA AGAGGGAGGT ATCAACAGAG ACTCCCCAAG AGCTAAGAGG AAACCACCTT GGACTGGAAG 000240
000241 TCAAGAGCCA AAATTCTAGA CTCTACTCTG TAATTAACTA CCTATTTGAA TTTGGAAAAA TCACCATCAA CTTTCCCAGC 000320
000321 CTCGTTCTCT GCATCTGGGA AATGAAGGCG TCGTCCAAAT GATTACAAGC TTCTTTTCCT GCTCTTATTG CATGATTCCA 000400
000401 CTTCCACAGC CCTCCAGCAT TTTTTAGCAG CTGCATCGCT CCATAGAGCC TGCAGAGGGC ACTAGACTGG GAATTAGAAA 000480
000481 ACCTGATTTC CCTTCCAGCT CCACCTCTGA CCAATTGCCT GACCCTGGTC AAATTGCTTA ACCTCTTCCT ATCTCAGCTC 000560
000561 CCTATCCATA AAACAGAGGG ACGAATAAAC TCTCCTCCTA CCACTAAGAG GTGTAGCCAG AGTTAATACC CTCATCGTCC 000640
000641 TTTGAGCTCA GCAGATGAAA GGCACTGAGA AAAGTACAAA GAATTTTTAT GTGCTATTGA CTTTATTTTA TTTTATGTGG 000720
000721 GGGAGGGAGC CGGCCCCAGC TGGAAAGCTG CTTTCTCTGA ATCAAAGGGC AGGAACCCAG CAAGTTTCTC AGGATTGGGG 000800
000801 CCTTAGACTG GGCTGTGTAT ACAGACAGTG CCAGCCAACC CCACAGTTCA GTTTCCTTTA ACCTGGTGCT CCAGGCAATA 000880
000881 ACTGTGCAAC TCTGCAATTT AACAATGTGT TCTTTGTCCC ACAACTGTTC TCGTTTCTCA ACTGCCCAGG TAATATGTTT 000960
000961 GGGCCTGTAG GAAGAGTCAA ATAGTTAATA AGGGAAGGGT TTGGCATGCC CTACGTAAGT TCTACCAGCA AGTCCCAACA 001040
001041 AGAAGGCATT CTGTGTCTCC TGATTCCTGA CCTACCCCCA AAATGTACAA ATGTACAAGG AATGAGCCCA CTTTCCCAGC 001120
001121 AGGCTGTAAT ACCAGTTTGG CCTATATCAA TGCATTGGTG AGCTGTGTTT TGTTTATGGT TTTATGCCAT CTATTTTCCC 001200
001201 ATGGATATTA TGTTTTCTAA AGAGCCCTTA AGTTTACGTC AGCTTTTAAA GCTACCAGCA GCACCATTTC AGTTCATATT 001280
001281 AAGCCCTTAA TATGGTATGA ATAGGAGAGC TATTAGACTA AAGAGCCATA ATCATCCCTG AGGAAAACAT CCATCACCAA 001360
001361 CATTTATGTG GTCCCTGAAC TTCTAAAAGG TGTCATCTCT CTGGGGTGTA TCTGGTGAGA GCTTTCTCTG GGAGATGCCA 001440
001441 AAAAGCCAAT GCATT