Difference between revisions of "ENST00000561978.1"
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | ''PCAT1'', a novel prostate cancer ncRNA alternately demonstrating either repression by PRC2 or an active role in promoting cell proliferation through transcriptional regulation of target genes. | |
==Annotated Information== | ==Annotated Information== | ||
| − | === | + | ===Name=== |
| − | + | ''PCAT1'':prostate cancer associated transcript 1 (non-protein coding) [https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:43022 (HGNC:43022)] | |
| + | |||
| + | ''PCA1'', ''PCAT-1''<ref name="ref1" /> | ||
| + | |||
| + | ===Characteristics=== | ||
| + | PCAT-1 resides in the 8q24 “gene desert” locus. PCAT-1 is located in the chromosome 8q24 gene desert approximately 725 kb upstream of the c-MYConcogene<ref name="ref1" />. | ||
===Function=== | ===Function=== | ||
| − | + | [[File:PCAT-1 is a marker of aggressive cancer and a PRC2-repressed ncRNA.jpg|right|thumb|400px|'''PCAT-1 is a marker of aggressive cancer and a PRC2-repressed ncRNA'''<ref name="ref1" />]] | |
| + | |||
| + | PCAT-1 biology may exhibit two distinct modalities: one in which PRC2 represses PCAT-1 and a second in which active PCAT-1 promotes cell proliferation. PCAT-1 and PRC2 may therefore characterize distinct subsets of prostate cancer<ref name="ref1" />. | ||
| + | |||
| + | PCAT-1 can stratify overall survival and correlated with distant metastasis in CRC patients, and the prognosis of CRC may be influenced by PCAT-1 expression, which suggested that PCAT-1 may be a potential diagnostic target in patients with colorectal cancer<ref name="ref2" />. | ||
===Regulation=== | ===Regulation=== | ||
| − | + | PCAT-1 was regulated by PRC2 complex in prostate cancer cells<ref name="ref1" />. | |
===Expression=== | ===Expression=== | ||
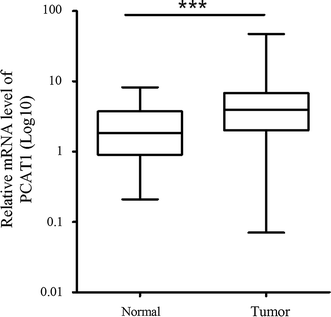
| − | + | PCAT-1, which is markedly overexpressed in a subset of prostate cancers, particularly metastases, and may contribute to cell proliferation in these tumors<ref name="ref1" /> | |
| − | = | + | PCAT-1 was overexpressed in colorectal cancer<ref name="ref2" />. |
| − | + | [[File:Comparison of PCAT-1 expression levels between CRC tumor tissues and matched normal tissues by real time PCR analysis.gif|right|thumb|400px|'''Comparison of PCAT-1 expression levels between CRC tumor tissues and matched normal tissues by real time PCR analysis'''<ref name="ref2" />]] | |
| − | === | + | siRNA sequences (in sense format) were as follows: |
| − | + | {|class='wikitable' style="text-align:center" | |
| + | |- | ||
| + | ! | PCAT-1 | ||
| + | ! | Sequence | ||
| + | |- | ||
| + | | | siRNA 1 | ||
| + | | | UUAAAGAGAUCCACAGUUAUU<ref name="ref1" /> | ||
| + | |- | ||
| + | | | siRNA 2 | ||
| + | | | GCAGAAACACCAAUGGAUAUU<ref name="ref1" /> | ||
| + | |- | ||
| + | | | siRNA 3 | ||
| + | | | AUACAUAAGACCAUGGAAAU<ref name="ref1" /> | ||
| + | |- | ||
| + | | | siRNA 4 | ||
| + | | | GAACCUAACUGGACUUUAAUU<ref name="ref1" /> | ||
| + | |- | ||
| + | ! | EZH2 | ||
| + | ! | Sequence | ||
| + | |- | ||
| + | | | siRNA | ||
| + | | | GAGGUUCAGACGAGCUGAUUU<ref name="ref1" /> | ||
| + | |} | ||
| − | + | ===Diseases=== | |
| + | * prostate cancer<ref name="ref1" /> | ||
| + | * colorectal cancer<ref name="ref2" /> | ||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
| − | + | * Michigan Center for Translational Pathology, University of Michigan Medical School, Ann Arbor, Michigan, USA<ref name="ref1" />. | |
| + | |||
| + | * State Key Laboratory of Oncology in South China, Sun Yat-Sen University Cancer Center, Guangzhou, China<ref name="ref2" />. | ||
==References== | ==References== | ||
| − | + | <references> | |
| + | <ref name="ref1">Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression[J]. Nature biotechnology. 2011,29(8):742-9.</ref>(1) | ||
| + | <ref name="ref2">Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, et al. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer[J]. Medical oncology. 2013,30(2):588.</ref>(2) | ||
| + | </references> | ||
{{basic| | {{basic| | ||
tID = ENST00000561978.1| | tID = ENST00000561978.1| | ||
source = Gencode19| | source = Gencode19| | ||
| − | same = lnc-POU5F1B-2:2,NONHSAT129015| | + | same = lnc-POU5F1B-2:2,NONHSAT129015,''PCAT1'',''PCAT-1'', ''PCA1''| |
classification = intergenic| | classification = intergenic| | ||
length = 1992 nt| | length = 1992 nt| | ||
Latest revision as of 09:48, 21 November 2018
PCAT1, a novel prostate cancer ncRNA alternately demonstrating either repression by PRC2 or an active role in promoting cell proliferation through transcriptional regulation of target genes.
Contents
Annotated Information
Name
PCAT1:prostate cancer associated transcript 1 (non-protein coding) (HGNC:43022)
PCA1, PCAT-1[1]
Characteristics
PCAT-1 resides in the 8q24 “gene desert” locus. PCAT-1 is located in the chromosome 8q24 gene desert approximately 725 kb upstream of the c-MYConcogene[1].
Function

PCAT-1 biology may exhibit two distinct modalities: one in which PRC2 represses PCAT-1 and a second in which active PCAT-1 promotes cell proliferation. PCAT-1 and PRC2 may therefore characterize distinct subsets of prostate cancer[1].
PCAT-1 can stratify overall survival and correlated with distant metastasis in CRC patients, and the prognosis of CRC may be influenced by PCAT-1 expression, which suggested that PCAT-1 may be a potential diagnostic target in patients with colorectal cancer[2].
Regulation
PCAT-1 was regulated by PRC2 complex in prostate cancer cells[1].
Expression
PCAT-1, which is markedly overexpressed in a subset of prostate cancers, particularly metastases, and may contribute to cell proliferation in these tumors[1]
PCAT-1 was overexpressed in colorectal cancer[2].
siRNA sequences (in sense format) were as follows:
| PCAT-1 | Sequence |
|---|---|
| siRNA 1 | UUAAAGAGAUCCACAGUUAUU[1] |
| siRNA 2 | GCAGAAACACCAAUGGAUAUU[1] |
| siRNA 3 | AUACAUAAGACCAUGGAAAU[1] |
| siRNA 4 | GAACCUAACUGGACUUUAAUU[1] |
| EZH2 | Sequence |
| siRNA | GAGGUUCAGACGAGCUGAUUU[1] |
Diseases
Labs working on this lncRNA
- Michigan Center for Translational Pathology, University of Michigan Medical School, Ann Arbor, Michigan, USA[1].
- State Key Laboratory of Oncology in South China, Sun Yat-Sen University Cancer Center, Guangzhou, China[2].
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression[J]. Nature biotechnology. 2011,29(8):742-9.
- ↑ 2.0 2.1 2.2 2.3 2.4 Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, et al. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer[J]. Medical oncology. 2013,30(2):588.
Basic Information
| Transcript ID |
ENST00000561978.1 |
| Source |
Gencode19 |
| Same with |
lnc-POU5F1B-2:2,NONHSAT129015,PCAT1,PCAT-1, PCA1 |
| Classification |
intergenic |
| Length |
1992 nt |
| Genomic location |
chr8+:128025399..128033259 |
| Exon number |
2 |
| Exons |
128025399..128026019,128031889..128033259 |
| Genome context |
|
| Sequence |
000001 ACACATGGAT ATTGGATATC TGCATAGGCA GCTTGCTCCA CGCCGGTGCC TACCTGTGCA GATGGGAAGG AAAGGAAAGT 000080 000081 GGCAAGGAGG CAGAGAAAGC ATCTGTACCC TTACAATTTG GTGAGACAAG AATGTATGAA TTCCCACAGG TCAAATTATA 000160 000161 ATGAAGAAAG GAACCTCTCT TGAGTACAAA GAGCTACCTA TGGTGGTCTG GAGCCGGAGG ACCACAGCAT CAAAGGATAT 000240 000241 AAGATGCATA GCCAACTGAG GAACCTGAGC AATTAAAGAG ATCCACAGTT AAGTCACACT TAACTGGCAC TTGTGGAAGC 000320 000321 CCCGCAAGGC CTGAAGGAGA GCTGACATAG GCACCCCAGA GAGCCAGAAT CTGGATCCCA TCTTAATAAG GCCATGAACA 000400 000401 CCAGTGGAGA AGAGGCAGAA ACACCAATGG ATAAGGAACA TTCACATCTT TCTTCCCATG TGCCTCTAAG TGCCAGTGCA 000480 000481 GGCCCCACAG GCCAAGCTAC AGGGAGAAAG GAGATGACGC AAAGGAACCT AACTGGACTT TAATCACTAG AAGTGAGAAG 000560 000561 AGAAATCTAT TGGAACCTCC CAAGATAATG CCAAGGGTCA AAGGGTGCGC AGATACATAA GACCATGGAA ATAATATCAG 000640 000641 ACAAAAAGCA GATTAGAGCA ATTTTCTTTT TCGAGTTCAA AATGGGTTAT AAAGCAGCGG AGACAAACCG CAACATCACC 000720 000721 AACGCCTTTG GCCCAGGAAC TGCTAATGAA GGTACAGTGC AGTCACTGTT CAGGAAGTTT TGCAAAGGAG ACTAGAGCCT 000800 000801 TGAAGATGAG GAGCATAGTG ACCAGCCATT GGAAGTCGAC AAAGACCAAT TGAGAGGAAT CATTGAAGCT GATCATCTTA 000880 000881 CAACTACACG AGAAGTTGTC AAAGAACGCA ATGTTGACCA TTGTGTGGTC TTTTCGCATT TGAAGCAAAT TGGAAAGGTG 000960 000961 AAAAACTTGA TAAGTGGGTG CCTTGTGAGC TCAGCAAAAA TCCAAAAAAA TAATCATTTT TAAGTGTTGT CTTCTCTTAT 001040 001041 TCTACGCAAC AACAATAACC ATTTTGCAAT CGGATTGTGA TGTGCAATGA AAAGTGGATT TGGGGCCGGG CGCGGTGGCT 001120 001121 CACGCCTGTA ATCTCAGCAC TTTGGAAGGC CAAGGCGGGC AGATCACGAG GTCAGGAGAT CAAGACCGTC CTGGCTAACA 001200 001201 CGGTGAAACC CCGTCTCTAC TGAAAATACA AAAAATTAGC CGGGTGTGGT GGCTGGCGCC TGTAGTCCCA GCTACAGGCT 001280 001281 GAGGCAGGAG AATGGCATGA ACCTGGGAGG CGGAGCTTGC AGTGAGCCGA GACCGT |