Difference between revisions of "PCAT7"
(→Expression) |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | ''PCAT7'': prostate cancer associated transcript 7 is associated with Prostate Cancer.<ref name="ref1"/> | ||
==Annotated Information== | ==Annotated Information== | ||
===Approved Symbol=== | ===Approved Symbol=== | ||
| − | PCAT7 | + | ''PCAT7'' [https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:48824 (HGNC:48824)] |
===Approved Name=== | ===Approved Name=== | ||
| − | prostate cancer associated transcript 7 (non-protein coding) | + | "prostate cancer associated transcript 7 (non-protein coding)" [https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:48824 (HGNC:48824)] |
| − | === | + | ===Alias=== |
| − | + | ''PCAN-R2'' : "prostate cancer-associated noncoding RNA 2" [https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:48824 (HGNC:48824)] | |
| − | |||
| − | PCAN-R2 | ||
| − | |||
| − | |||
===RefSeq ID=== | ===RefSeq ID=== | ||
NR_121566 | NR_121566 | ||
===Ensembl ID=== | ===Ensembl ID=== | ||
ENSG00000231806 | ENSG00000231806 | ||
| − | === | + | |
| − | + | ===Characteristics=== | |
| − | ===Sequence | + | [[File:PCAT7.jpg|right|thumb|400px|'''Functional validation of PCAN-R1 and PCAN-R2 |
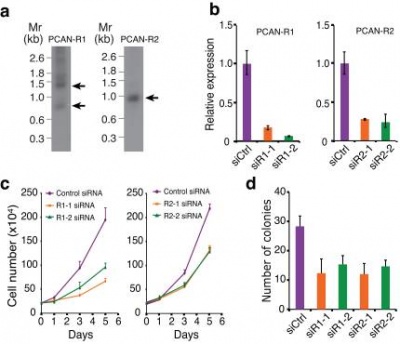
| + | (a) The Northern blot of PCAN-R1 and PCAN-R2 transcripts was shown (Mr: RNA marker). (b) The relative expression level of PCAN-R1 and PCAN-R2 upon knockdown by two different siRNA (orange and green) and upon control siRNA treatment (purple) was shown. (c) The growth curves of LNCaP cell with or without targeted siRNA-mediated knockdown of PCAN-R1 or PCAN-R2 were shown. The growth curves of control siRNA-treated cells and the growth curves of two targeted siRNA-treated cells were plotted in purple, orange, and green, respectively. Data were shown as Mean+S.D. n=3. (d) The number of soft-agar colony formation of LNCaP cell with or without targeted siRNA-mediated knockdown of PCAN-R1 or PCAN-R2 was shown.'''<ref name="ref1" />]] | ||
| + | |||
| + | ''PCAT7'' is a 1937-bp long non coding RNA gene located at chromosome 9q22.32. Studies suggest that PCAT7 might be acted as oncogene, a potential diagnostic marker, and a novel therapeutic target for cancer. <ref name="ref2"/> | ||
| + | |||
| + | ===Function=== | ||
| + | In nasopharyngeal carcinoma NPC tumorigenesis, ''PCAT7'' acts as a key regulator through regulating miR-134-5p/ELF2 signal pathway. It mediates its oncogenic role by functioning as a competitive endogenous RNA (ceRNA) for miR-134-5p and directly interacts with miR-134-5p at recognized sites. ''PCAT7'' accelerates NPC cell growth partially by spongeing miR-134-5p, releasing the miR-134-5p's inhibitory role for its target gene ELF2 therefore up-regulating ELF2 and subsequently initiating ELF2 signaling pathway. <ref name="ref2"/> | ||
| + | |||
| + | ===Expression=== | ||
| + | ''PCAN-R2'' or ''PCAT7'' is upregulated in metastatic prostate cancer and knockdown of this lncRNA resulted in substantial decrease in both cell growth and soft-agar colony formation in the androgen-dependent prostate cancer cell line LNCaP. <ref name="ref1"/> | ||
| + | |||
| + | ''PCAT7'' is higher expressed in NPC tissues than corresponding noncancerous tissues, and higher ''PCAT7'' expression is associated with worse survival in NPC patients. Silencing ''PCAT7'' exerted tumor-suppressive roles by suppressing NPC cell proliferation in vitro and tumor growth in vivo. <ref name="ref2"/> | ||
| + | |||
| + | ''PCAT7'' was found to be overexpressed in Non-Small-Cell Lung Cancer (NSCLC) tissues (compared with corresponding non-tumor tissues) and NSCLC cells (compared with normal cell line 16-HBE). Overexpression of ''PCAT7'' resulted in the promotion of tumor cell proliferation, inhibition of cells apoptosis, facilitation of cells metastasis, and formation of EMT (epithelial-to-mesenchymal transition) phenotype, while ''PCAT7'' expression deletion remarkably prohibited cell proliferation, accelerated their apoptosis, weakened metastasis, and reversed EMT to MET. <ref name="ref3"/> | ||
| + | {| class='wikitable' style="text-align:center" | ||
| + | |- | ||
| + | ! | Experiment | ||
| + | ! | Forward primer | ||
| + | ! | Reverse primer | ||
| + | |- | ||
| + | | rowspan="1"|quantitative real-time PCR | ||
| + | | | 5'-AAACAAGCCAACCGCACAAT- 3' | ||
| + | | | 5'-CCTGCTTGCTGTGTTACTGC- 3'<ref name="ref2" /> | ||
| + | |} | ||
| + | |||
| + | ===Disease=== | ||
| + | * Nasopharyngeal carcinoma (NPC) <ref name="ref2"/> | ||
| + | * Non-Small-Cell Lung (NSCLC) <ref name="ref3"/> | ||
| + | * Prostate Cancer <ref name="ref1"/> | ||
| + | |||
| + | ==Labs working on this lncRNA== | ||
| + | * Department of Bioinformatics, School of Life Sciences and Technology, Tongji University, Shanghai, China | ||
| + | * Department of Otolaryngology-Head and Neck Surgery, Renmin Hospital of Wuhan University, Wuhan 430060, Hubei Province, People’s Republic of China | ||
| + | * Department of Thoracic Surgery, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P.R. China | ||
| + | |||
| + | ==Sequence== | ||
>gi|651169114|ref|NR_121566.1| Homo sapiens prostate cancer associated transcript 7 (non-protein coding) (PCAT7), transcript variant 1, long non-coding RNA | >gi|651169114|ref|NR_121566.1| Homo sapiens prostate cancer associated transcript 7 (non-protein coding) (PCAT7), transcript variant 1, long non-coding RNA | ||
<dnaseq>AGAACATGCAGTCTAGGAACCGGCATGCGCATAACCTCAGGATATAAATAATGCTGAAGCAGAGTTACGTTTTTTTTGTTGTTGTTTTTTTTGTTTTTGTTTTTTTAGGTTTCCGTGTGTTTCTATTGAGCTGCTCAGTGCCCGGCTTAGAAGACCAGGAAAAGGAGTCACAGGTCGTATGCTGGAGGCTTGAGCCGCGGCACCGTGGCGCGGCTCGCCTCGCTGCGGTTGGTGGTGGCGGTGGACATTGCAGCGCGGCTGGAGGGGTAGGAAGAAGAAGATAGTTTTGGGATAGATAGAGGGTCCTTAGACAAGGTGCAAGACAAACAGAAGAGGGCATGTGGGGTCAAACTCGCTAGCTGCCTGCCTGATTTTTCTGCACACAGGTGAGATATTCCTGCACATCCTCTGGTGACCCCAGAATGAGGGGGACTCGCTGGTGAATTGCCTCGGGCTTCACGTCCAGTACAGGCTGGGTCCCCGTGGTCGCCAAGCCTCCTGCCTGCTCAATGATGTAGGCCACGGGATTGCATTCATACAGGAGCCGGAGCTGTGGAGGAACAGAGGCAGGACAAATTCACCAAGAGCCTAGCAACATGAAGAGAGATGCCAGGAAGAAGAGAGAAGCCAGGAAACAAGCCAACCGCACAATCCCCACATCAGAGCAGGAGAAGATGGGGGCCTGCTGGCAGAGCTGGGGCTTGGCTGTGGTCACTCTGAACCTGCTCTTTGGTGTTTTCATGAGTGGTGGGAAGAATAGGGACCATATGGAGCCCACACAGGAAGCTCTAGCAGTAACACAGCAAGCAGGAAGACAATTCTAAGGAAGCAGCCCATAGTCTTCTTTCTTTTCCTGTGCATCTTCCACTGTCAGTGAGGCTCCTCATTTATGGTGAACCCAACTGTGTGTATCTCCCAAGTTCTCACCCGCAGATTAATGTTTTCAGGAAGATAGGCCATCAACAGTGAGAGGAAGAAGTTACATTGTCGTATGAGGGATGCATTTTAACCATTAATTTGTGGTACAGGCTGGGCGCAGTGGCTTATGCATGTAATCCCAGCACTTTGGGAGGCCGAGGTGGGTGGATCACGAGGTCAGGAGATCGAGACCATCCTGGCTAACATGGTGAAACCCCGTCTTTACTAAATATACAAAAAATTGGCCGGGCGTGGTGGTGGGCACCTGTAGTCCCAGCTACTCGGGGGGCTGAGGCAGGAGAATGGTGTGAACCCGGGAGGCAGAGCTTGCAGTGAGCCGAGATCGCGCCACTGCACTCCAGCCTGGATGACAGAGCAAGACTCCATCTCAAAAAAAAAAAAAA</dnaseq> | <dnaseq>AGAACATGCAGTCTAGGAACCGGCATGCGCATAACCTCAGGATATAAATAATGCTGAAGCAGAGTTACGTTTTTTTTGTTGTTGTTTTTTTTGTTTTTGTTTTTTTAGGTTTCCGTGTGTTTCTATTGAGCTGCTCAGTGCCCGGCTTAGAAGACCAGGAAAAGGAGTCACAGGTCGTATGCTGGAGGCTTGAGCCGCGGCACCGTGGCGCGGCTCGCCTCGCTGCGGTTGGTGGTGGCGGTGGACATTGCAGCGCGGCTGGAGGGGTAGGAAGAAGAAGATAGTTTTGGGATAGATAGAGGGTCCTTAGACAAGGTGCAAGACAAACAGAAGAGGGCATGTGGGGTCAAACTCGCTAGCTGCCTGCCTGATTTTTCTGCACACAGGTGAGATATTCCTGCACATCCTCTGGTGACCCCAGAATGAGGGGGACTCGCTGGTGAATTGCCTCGGGCTTCACGTCCAGTACAGGCTGGGTCCCCGTGGTCGCCAAGCCTCCTGCCTGCTCAATGATGTAGGCCACGGGATTGCATTCATACAGGAGCCGGAGCTGTGGAGGAACAGAGGCAGGACAAATTCACCAAGAGCCTAGCAACATGAAGAGAGATGCCAGGAAGAAGAGAGAAGCCAGGAAACAAGCCAACCGCACAATCCCCACATCAGAGCAGGAGAAGATGGGGGCCTGCTGGCAGAGCTGGGGCTTGGCTGTGGTCACTCTGAACCTGCTCTTTGGTGTTTTCATGAGTGGTGGGAAGAATAGGGACCATATGGAGCCCACACAGGAAGCTCTAGCAGTAACACAGCAAGCAGGAAGACAATTCTAAGGAAGCAGCCCATAGTCTTCTTTCTTTTCCTGTGCATCTTCCACTGTCAGTGAGGCTCCTCATTTATGGTGAACCCAACTGTGTGTATCTCCCAAGTTCTCACCCGCAGATTAATGTTTTCAGGAAGATAGGCCATCAACAGTGAGAGGAAGAAGTTACATTGTCGTATGAGGGATGCATTTTAACCATTAATTTGTGGTACAGGCTGGGCGCAGTGGCTTATGCATGTAATCCCAGCACTTTGGGAGGCCGAGGTGGGTGGATCACGAGGTCAGGAGATCGAGACCATCCTGGCTAACATGGTGAAACCCCGTCTTTACTAAATATACAAAAAATTGGCCGGGCGTGGTGGTGGGCACCTGTAGTCCCAGCTACTCGGGGGGCTGAGGCAGGAGAATGGTGTGAACCCGGGAGGCAGAGCTTGCAGTGAGCCGAGATCGCGCCACTGCACTCCAGCCTGGATGACAGAGCAAGACTCCATCTCAAAAAAAAAAAAAA</dnaseq> | ||
| + | |||
| + | ==References== | ||
| + | <references> | ||
| + | <ref name="ref1"> Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer[J]. Nature structural & molecular biology. 2013,20(7):908-13.</ref>(1) | ||
| + | |||
| + | <ref name="ref2"> Liu Y, Tao Z, Qu J, Zhou X & Zhang C. Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma[J]. Biochemical and biophysical research communications. 2017, 491(2):374-381.</ref>(2) | ||
| + | |||
| + | <ref name="ref3"> Liu Q, Wu Y, Xiao J & Zou J. Long Non-Coding RNA Prostate Cancer-Associated Transcript 7 (PCAT7) Induces Poor Prognosis and Promotes Tumorigenesis by Inhibiting mir-134-5p in Non-Small-Cell Lung (NSCLC)[J]. Medical science monitor : international medical journal of experimental and clinical research. 2017, 23:6089-6098.</ref>(3) | ||
| + | </references> | ||
Latest revision as of 08:00, 23 November 2018
PCAT7: prostate cancer associated transcript 7 is associated with Prostate Cancer.[1]
Contents
Annotated Information
Approved Symbol
PCAT7 (HGNC:48824)
Approved Name
"prostate cancer associated transcript 7 (non-protein coding)" (HGNC:48824)
Alias
PCAN-R2 : "prostate cancer-associated noncoding RNA 2" (HGNC:48824)
RefSeq ID
NR_121566
Ensembl ID
ENSG00000231806
Characteristics
PCAT7 is a 1937-bp long non coding RNA gene located at chromosome 9q22.32. Studies suggest that PCAT7 might be acted as oncogene, a potential diagnostic marker, and a novel therapeutic target for cancer. [2]
Function
In nasopharyngeal carcinoma NPC tumorigenesis, PCAT7 acts as a key regulator through regulating miR-134-5p/ELF2 signal pathway. It mediates its oncogenic role by functioning as a competitive endogenous RNA (ceRNA) for miR-134-5p and directly interacts with miR-134-5p at recognized sites. PCAT7 accelerates NPC cell growth partially by spongeing miR-134-5p, releasing the miR-134-5p's inhibitory role for its target gene ELF2 therefore up-regulating ELF2 and subsequently initiating ELF2 signaling pathway. [2]
Expression
PCAN-R2 or PCAT7 is upregulated in metastatic prostate cancer and knockdown of this lncRNA resulted in substantial decrease in both cell growth and soft-agar colony formation in the androgen-dependent prostate cancer cell line LNCaP. [1]
PCAT7 is higher expressed in NPC tissues than corresponding noncancerous tissues, and higher PCAT7 expression is associated with worse survival in NPC patients. Silencing PCAT7 exerted tumor-suppressive roles by suppressing NPC cell proliferation in vitro and tumor growth in vivo. [2]
PCAT7 was found to be overexpressed in Non-Small-Cell Lung Cancer (NSCLC) tissues (compared with corresponding non-tumor tissues) and NSCLC cells (compared with normal cell line 16-HBE). Overexpression of PCAT7 resulted in the promotion of tumor cell proliferation, inhibition of cells apoptosis, facilitation of cells metastasis, and formation of EMT (epithelial-to-mesenchymal transition) phenotype, while PCAT7 expression deletion remarkably prohibited cell proliferation, accelerated their apoptosis, weakened metastasis, and reversed EMT to MET. [3]
| Experiment | Forward primer | Reverse primer |
|---|---|---|
| quantitative real-time PCR | 5'-AAACAAGCCAACCGCACAAT- 3' | 5'-CCTGCTTGCTGTGTTACTGC- 3'[2] |
Disease
Labs working on this lncRNA
- Department of Bioinformatics, School of Life Sciences and Technology, Tongji University, Shanghai, China
- Department of Otolaryngology-Head and Neck Surgery, Renmin Hospital of Wuhan University, Wuhan 430060, Hubei Province, People’s Republic of China
- Department of Thoracic Surgery, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P.R. China
Sequence
>gi|651169114|ref|NR_121566.1| Homo sapiens prostate cancer associated transcript 7 (non-protein coding) (PCAT7), transcript variant 1, long non-coding RNA
000081 GTTGTTTTTT TTGTTTTTGT TTTTTTAGGT TTCCGTGTGT TTCTATTGAG CTGCTCAGTG CCCGGCTTAG AAGACCAGGA 000160
000161 AAAGGAGTCA CAGGTCGTAT GCTGGAGGCT TGAGCCGCGG CACCGTGGCG CGGCTCGCCT CGCTGCGGTT GGTGGTGGCG 000240
000241 GTGGACATTG CAGCGCGGCT GGAGGGGTAG GAAGAAGAAG ATAGTTTTGG GATAGATAGA GGGTCCTTAG ACAAGGTGCA 000320
000321 AGACAAACAG AAGAGGGCAT GTGGGGTCAA ACTCGCTAGC TGCCTGCCTG ATTTTTCTGC ACACAGGTGA GATATTCCTG 000400
000401 CACATCCTCT GGTGACCCCA GAATGAGGGG GACTCGCTGG TGAATTGCCT CGGGCTTCAC GTCCAGTACA GGCTGGGTCC 000480
000481 CCGTGGTCGC CAAGCCTCCT GCCTGCTCAA TGATGTAGGC CACGGGATTG CATTCATACA GGAGCCGGAG CTGTGGAGGA 000560
000561 ACAGAGGCAG GACAAATTCA CCAAGAGCCT AGCAACATGA AGAGAGATGC CAGGAAGAAG AGAGAAGCCA GGAAACAAGC 000640
000641 CAACCGCACA ATCCCCACAT CAGAGCAGGA GAAGATGGGG GCCTGCTGGC AGAGCTGGGG CTTGGCTGTG GTCACTCTGA 000720
000721 ACCTGCTCTT TGGTGTTTTC ATGAGTGGTG GGAAGAATAG GGACCATATG GAGCCCACAC AGGAAGCTCT AGCAGTAACA 000800
000801 CAGCAAGCAG GAAGACAATT CTAAGGAAGC AGCCCATAGT CTTCTTTCTT TTCCTGTGCA TCTTCCACTG TCAGTGAGGC 000880
000881 TCCTCATTTA TGGTGAACCC AACTGTGTGT ATCTCCCAAG TTCTCACCCG CAGATTAATG TTTTCAGGAA GATAGGCCAT 000960
000961 CAACAGTGAG AGGAAGAAGT TACATTGTCG TATGAGGGAT GCATTTTAAC CATTAATTTG TGGTACAGGC TGGGCGCAGT 001040
001041 GGCTTATGCA TGTAATCCCA GCACTTTGGG AGGCCGAGGT GGGTGGATCA CGAGGTCAGG AGATCGAGAC CATCCTGGCT 001120
001121 AACATGGTGA AACCCCGTCT TTACTAAATA TACAAAAAAT TGGCCGGGCG TGGTGGTGGG CACCTGTAGT CCCAGCTACT 001200
001201 CGGGGGGCTG AGGCAGGAGA ATGGTGTGAA CCCGGGAGGC AGAGCTTGCA GTGAGCCGAG ATCGCGCCAC TGCACTCCAG 001280
001281 CCTGGATGAC AGAGCAAGAC TCCATCTCAA AAAAAAAAAA AA
References
- ↑ 1.0 1.1 1.2 1.3 Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer[J]. Nature structural & molecular biology. 2013,20(7):908-13.
- ↑ 2.0 2.1 2.2 2.3 2.4 Liu Y, Tao Z, Qu J, Zhou X & Zhang C. Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma[J]. Biochemical and biophysical research communications. 2017, 491(2):374-381.
- ↑ 3.0 3.1 Liu Q, Wu Y, Xiao J & Zou J. Long Non-Coding RNA Prostate Cancer-Associated Transcript 7 (PCAT7) Induces Poor Prognosis and Promotes Tumorigenesis by Inhibiting mir-134-5p in Non-Small-Cell Lung (NSCLC)[J]. Medical science monitor : international medical journal of experimental and clinical research. 2017, 23:6089-6098.