Difference between revisions of "ENST00000416928.2"
| (One intermediate revision by the same user not shown) | |||
| Line 7: | Line 7: | ||
"HOXD cluster antisense RNA 1", "HOXD cluster antisense RNA 1 (non-protein coding)", ''HOXD-AS1''<ref name="ref2" /> | "HOXD cluster antisense RNA 1", "HOXD cluster antisense RNA 1 (non-protein coding)", ''HOXD-AS1''<ref name="ref2" /> | ||
| + | |||
| + | LncBook ID: [https://bigd.big.ac.cn/lncbook/transcript?transid=HSALNT0043560 HSALNT0043560] | ||
===Function=== | ===Function=== | ||
| Line 17: | Line 19: | ||
===Diseases=== | ===Diseases=== | ||
| − | + | neuroblastoma<ref name="ref2" /> | |
===Expression=== | ===Expression=== | ||
Latest revision as of 13:11, 11 August 2019
HAGLR, which is induced by RA and regulated via PI3K/Akt pathway, controls genes involved in RA signaling, angiogenesis and inflammation.
Contents
Annotated Information
Name
HAGLR:HOXD antisense growth-associated long non-coding RNA(HGNC nomenclature)
Mdgt[1]
"HOXD cluster antisense RNA 1", "HOXD cluster antisense RNA 1 (non-protein coding)", HOXD-AS1[2]
LncBook ID: HSALNT0043560
Function

HOXD-AS1 is evolutionary conserved among hominids and has all bona fide features of a gene. HOXD-AS1 is a subject to morphogenic regulation, is activated by PI3K/Akt pathway and itself is involved in control of RA-induced cell differentiation. Knock-down experiments revealed that HOXD-AS1 controls expression levels of clinically significant protein-coding genes involved in angiogenesis and inflammation, the hallmarks of metastatic cancer[2].
Regulation
HOXD-AS1 is regulated by PI3K/Akt pathway[2].
Diseases
neuroblastoma[2]
Expression
Mdgt is highly expressed in testes, and more moderately in brain, kidney, colon and skeletal muscles[1].
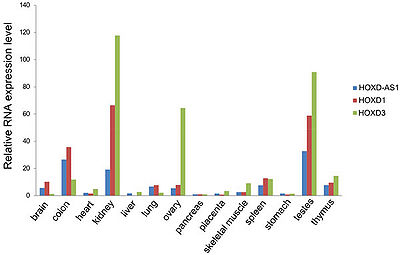
HOXD-AS1 is expressed in multiple tissues with the highest levels of expression in kidney, colon and testes and the lowest in liver, heart, pancreas and stomach. In general, HOXD-AS1 demonstrated highly correlated expression with its neighboring protein coding genes HOXD1 and HOXD3, thus implying that they are subject to the common regulatory mechanism[2].
Evolution
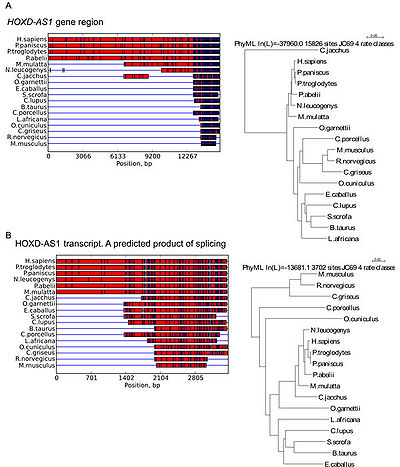
More than 96% of this gene sequence is evolutionary conserved among hominids (H. sapiens, P. paniscus, P. troglodytes and P. abelii). At the same time, only 12% to 52% of HOXD-AS1sequence is shared with other primates (M. mulatta, N. leucogenys, C. jacchus and O. garnettii), comparable to 11% of dog (C. lupus) and 8-12% of ungulates (E. caballus, S. scrofa and B. taurus). Thus, a gap between HOXD-AS1 genomic sequence in apes and other monkeys is apparent[2].
Cellular Localization
HOXD-AS1 is evenly distributed between nucleus and cytoplasm[2].
Labs working on this lncRNA
Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, United States[1].
Department of Genome and Gene Expression Data Analysis, Bioinformatics Institute, Agency for Science, Technology and Research (A*STAR), Matrix, 138671 Singapore[2].
References
- ↑ 1.0 1.1 1.2 Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development[J]. eLife. 2013,2:e01749.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 Yarmishyn AA, Batagov AO, Tan JZ, Sundaram GM, Sampath P, Kuznetsov VA, et al. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome[J]. BMC genomics. 2014,15 Suppl 9:S7.
Basic Information
| Transcript ID |
ENST00000416928.2 |
| Source |
Gencode19 |
| Same with |
lnc-AC009336.1-2:6,HAGLR,Mdgt,HOXD-AS1 |
| Classification |
intergenic |
| Length |
4235 nt |
| Genomic location |
chr2-:177037923..177043012 |
| Exon number |
3 |
| Exons |
177037923..177041420,177042060..177042222,177042439..177043012 |
| Genome context |
|
| Sequence |
000001 CGGCCGCCAG CCGGTAGGGT GATTAACTCA CCGTCTGTCT TTGCAAAAGT GTTCCAGGTG GATCAGGAGC GCCAGCCCAC 000080 000081 CCCGCCCGCA GCCTGGCCGC TCGCTGAGGC GCGGAGAGCG CGGAACGAGC GCGCGCATCA CGTGGCGGCG CGGACGTTTG 000160 000161 TGCCGCGCGC CCGCCAGACC GCCCAGGAGG CGGCGGCTAT TGTTCCCCGC AGGGGCCCGG GGCGCCCTGA AACGGGGTCC 000240 000241 ACGTCAGCCC GCTCCCGAGC GTGTCCTCAG GTCTAAGGAC GGGTGGGGCT TCCTTTGTGT GTTCTCTGGA AGGGGCTGGG 000320 000321 ATGGCGTCGT TGCTGCCCCA AGGGGTAACC GAGGGGGATT GAGGGAAGAT GGTACCAGCT CCCTAGAGCT CAGCTCACCA 000400 000401 GACCTACTCT TCCGCTCGAC CCTTGCAGAG GAGGTGGGGA CGGATATGGT ATTTGTAGGG CAGCGCTGTG AACAATGGAG 000480 000481 CCATCAAAAA GGCCTTCTCC CAGCCTCCTC ACCAACCACG GACATTCCTG GCCCAACAAG ACCTCCTGGC CAGGTTTTGA 000560 000561 GCCCTGCTTC CCAGATCTGG GGACCCTTCA CCTGCCTCTA CTACTGCAAA TGATTGGATA TTCGTCTGAC TTGGCTCTTC 000640 000641 CCTAATGTGT GGAACTAATT TTGTTCACTC TGTTTCATGC TGGACCTGGC CTGAAAATGG GCCCTTATAT TGTCTTTGCA 000720 000721 CTTTCCAACT TGCCAAGTTT CTGAAAGAAG GACCAAAGTA ATGCCAAGAA CTCCCAGATG AAGAGGACCT TCCTTAGACC 000800 000801 TATAACAGAT TCACTACTTG ACTTCAGAGG GAAGTTTCAG CTAATTCTCA GGAAAAGGTC AAAACAGGCC TGTGACTGTC 000880 000881 CCTTGAATAC TGAAGGTATA AACAAGCTTC TGGCTCCTCC AGGTCAAGCA GAGGAACACT TTTATTAGAA TGAAGGGTTA 000960 000961 CTTCTTTGTT ATTCAAACAC ACACCCACAC ACATCCTCTG TAGTCTCCCT AATGAAAAAC CTCAACAGAT GGAGAGCCTG 001040 001041 AGCTCCCAAG TCCGCAGGCT GAGAAGATGC ACTTCACTCA AGGGCCAGTG GGCTGGTACA GACTAGGGAA GAGAAAAGGG 001120 001121 ACAAAGGCTG CTAGTATAGG TAATCACTAT CTTTTAATAC TCCTAACACA TAGAAATTCG AAAAATCTTG TTCAGCAATG 001200 001201 TTAGCAGCCT GTGAGCCTCG GAAACTAATT TGGCTGATGT GTCTCTGATA GGGAGCTTGG GGCTGCACCA AGGGGAAACA 001280 001281 TTCTCCTCCT GTATTCTCTC CTCTCTCTCG TTTGAAAGCC TGTACAAAGA AATGGTACAA ATGGATGGTT AACTTAATGA 001360 001361 TTCCATCAGA GGCAGATTTT TCTCTGTCAG GTCCCTGGAT AAGGTGGAGG TCTGAAAGCC AGGAGACAGA ATATGTACTT 001440 001441 AATCGCCCTT TCTGACCTGC TTACCAGTGA CCAGGACTAC TCTCCAGGCC TTTCTTCTGC CACTTCCCCA TTCTTCCTGG 001520 001521 CTAGTTCAGT TGGGTCTCAG CACTTCAACT CCTGGCTGGA TCCTGAGTTT TTCTTGGTTG CAGGGAAAGC TCTCACTGTC 001600 001601 TGCATCTCCT TCCCCACCCC CACCTCCCGT TTTGCAGCTG GCCATTCTGG CTGCCTCACA GCTATCCTGC CTTAATTCAT 001680 001681 ATTTGATTGC CTGGACAGAA CCACCTAGAA TGTCCAGACG ACTGCCACCC CTCAGCCATG GGCCTGAGAT CCTGCCCTGA 001760 001761 CCCCTTTCTT CAGACCCCAG GAGGACCCAC AATCTCCCTG ATCCCCACCT TCCCCAAAGT AGTCTCTAGG AAAAAAGCAG 001840 001841 AGCTGGCTAA GGCCTGAGAC AGAGCCATTT CTTCCCCTTG GGCAAACGAG GTAGACCCTC CCAGAGACCA ATGCCGGAGA 001920 001921 GGCCCGGATC CGAGCTCCCT TTATATGGCC TCCCCCATCA CGGATCCCGT GGGAAAACCT GACACACAAT GGCGCGGCTG 002000 002001 CTCCGGGCAC CCTCAACCCA GCGCCCTGCG GCAAGGCCTG GAGCAGCCTG GTGGTCTTGG TGCGCTCCCT TTGCCCTCCC 002080 002081 CGCAACCCTC TGCGAGCACT CGTCCCAGGA GAGAGACCCT GGCTAGGGTG GAGGTTGCCT CCCGCAAAGG GCGCTGTTGC 002160 002161 TGGGGCCTCC CCTCCCCCAT TCTCTTGTTT TTCTGCTTCT GAGACTCGGG AGGGAGGCGG CAGGCGCCTC CGCAGCCGAT 002240 002241 AGAGCAGCTA TTGTGACCTT CCCTTGCAGA GCGGGAACCC AGCCGAGTGT GTACCGCCGG CGGTGGCGGG GGAGGAGGCG 002320 002321 ACTGTGTGTG CAAACCTCAG TCGGAGAGGG GGTGGCGCTG GGAAATAAAG GTTTGCAACC TCAGGCAGTT GGGCCCTTCA 002400 002401 GAGGGCGTTT GGTTGTGTTT AGACTCGGTT GCTTTCTAAC ACTAATTGCA GGCTCAAAAG GAGCGGAATT TATTGCGAAG 002480 002481 GCCAAGGCGG CTTGTTTATA ATTTATGAAG TTTTAAATGG CTGTGCCGCG GGCCCCAACT TTCTCCTGGG CTGGCAGCCG 002560 002561 CGCGGAAACG CGCCCTCCCC CGGCTCGCCG CCCACCCCCA CCCACGCCCC TCAGATGAGT CTCACGCCAA TGGCAGGAAC 002640 002641 AGATCCGAGC TATTAGAAGT CTCGGGAACC TCCAGAGATG ACAGAAGGTG GCAGATTTGG TCCAAGCCCT CACCCCACTC 002720 002721 GAGAGGAGGG CGTCGGGGAA AGCTGTGGCT GCCTGGAGAG GGAGCCCCTG ATCCCACACT TCCTTGGAGC CTGAGTGTGA 002800 002801 GAGGAAGCTA ATGTCAGCCG GGCTAGACAC CGTTTAAAGT CTAATCCATG AAACCCTAAG CCGCAAATGA CACTTGGGGC 002880 002881 TTTGCAGACA CCCGCAGACT CAGCAGATAC TCCTGCGGTA ACACACTGTG CTGGAAAACG CCTGGAGCCA TGACACATTA 002960 002961 CGTATTTTTA TTATTATTTA TGGTAGGAAG AGGAGATGAG GGAAAATCGG GCTGAATCTA AGCAGCCTGA CAATTCAGCT 003040 003041 AGTTAATATT TAGCCAGTTA AGAAAGGTGG TTGTAATCCC TGCAAGGCAG TGAGCTCTCC TTTGATTTCT CTAATAGCTT 003120 003121 AAAAAATAAT TTCTTACAAG ACAATTAGAA ATTCTGCCTA TGGATGCAAA TTTCATAGAA AACAATGTTA AGCCTTGAAA 003200 003201 GTGGGTAAAA TGTGCCTTCG ATTGAAGTCA TTTATTCTGT TGATACACTT CTACAACATA TATATTTTAA ACCTGCTCAG 003280 003281 ATAAAGTGAA ATTTAGCAAA AGGATATCCC CTCCCACCCC CAAATCTAAC ATGCCATTTT AAAAACAGCA CAAAGGAACA 003360 003361 AGGAAACTAA ATATTATTTT GAGCCATCAT TACTTAGTGA AATACAAACA TCCAAATAGA AACGATTGTT GTTCCACCAG 003440 003441 AAGATTAGAA GTTAAATTAT TTCCACACAA TAGGAATTTC ACTTCGTGTC ATCGGGGATG TAGTAGGAGT TGCATTATTT 003520 003521 GTCAAACCAA ATAGAGATGC GTGGGCTGAC TTTGCTGAAA TACAAAAGAA TTGAACCTTA TCTGAAGGCG TTTTCTAAGT 003600 003601 GGCCCCTCCC ATAGAAGCCG GGAAGTTGTC CACCCCTAGC AGAAGCCACA TTAATATTTA CAAAATGGTT ATAAATCCTT 003680 003681 GAAGACATTT TTGCCCATTT TGTATAAGAA ACAGGAGTAA ACAAGGTGTT TATACTTGTT ATGTTTCATT AATGAAATAT 003760 003761 TATTACTTTA CATCTTGGGG AATGTTTGTT TTCTTTAGAA ATGAACTAAT GATCACTACA AA |