Difference between revisions of "MROCKI"
(Created page with "MARCKS (Regulator of Cytokines and Inflammation), an new gliomagenesis-specific candidate lncRNAs involved in epigenetic regulation, which was induced by multiple TLR stimuli...") |
(→Name) |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
Approved name:MARCKS cis regulating lncRNA promoter of cytokines and inflammation | Approved name:MARCKS cis regulating lncRNA promoter of cytokines and inflammation | ||
| − | HGNC | + | HGNC ID:50323 |
Previous names:long intergenic non-protein coding RNA 1268 | Previous names:long intergenic non-protein coding RNA 1268 | ||
| Line 15: | Line 15: | ||
RefSeq ID:NR_038863 | RefSeq ID:NR_038863 | ||
| + | |||
===Characteristics=== | ===Characteristics=== | ||
MARCKS harbors an intragenic CpG island and is closely flanked by the uncharacterized MROCKI(hg19: chr6:114,189,179-114,194,512), | MARCKS harbors an intragenic CpG island and is closely flanked by the uncharacterized MROCKI(hg19: chr6:114,189,179-114,194,512), | ||
| Line 27: | Line 28: | ||
MROCKI interacted with APEX1 (apurinic/apyrimidinic endodeoxyribonuclease 1) to form a ribonucleoprotein complex at the MARCKS promoter. <ref name="ref3" /> MROCKI-APEX1 recruited the histone deacetylase HDAC1, which removed the H3K27ac modification from the promoter, thus reducing MARCKS transcription and subsequent Ca2+ signaling and inflammatory gene expression.<ref name="ref3" /> | MROCKI interacted with APEX1 (apurinic/apyrimidinic endodeoxyribonuclease 1) to form a ribonucleoprotein complex at the MARCKS promoter. <ref name="ref3" /> MROCKI-APEX1 recruited the histone deacetylase HDAC1, which removed the H3K27ac modification from the promoter, thus reducing MARCKS transcription and subsequent Ca2+ signaling and inflammatory gene expression.<ref name="ref3" /> | ||
===Expression=== | ===Expression=== | ||
| − | [[File: | + | [[File: MORICA.jpg|thumb|300px| Expression levels were significantly increased in 105/125 samples compared to brain reference RNA (A), and all glioma subtypes showed positive ΔΔCT average values(B).<ref name="ref2"/>]] |
In all glioma subtypes levels of MROCKI were significantly higher in comparison to normal brain reference RNA, and expression was inversely associated with promoter methylation.<ref name="ref2" /> | In all glioma subtypes levels of MROCKI were significantly higher in comparison to normal brain reference RNA, and expression was inversely associated with promoter methylation.<ref name="ref2" /> | ||
MROCKI expression is significantly increased in violent suicides irrespective of MARCKS transcription levels.<ref name="ref1" /> | MROCKI expression is significantly increased in violent suicides irrespective of MARCKS transcription levels.<ref name="ref1" /> | ||
| + | |||
===Diseases=== | ===Diseases=== | ||
Glioma tumours<ref name="ref2" /> | Glioma tumours<ref name="ref2" /> | ||
Latest revision as of 02:45, 19 February 2021
MARCKS (Regulator of Cytokines and Inflammation), an new gliomagenesis-specific candidate lncRNAs involved in epigenetic regulation, which was induced by multiple TLR stimuli and acted as a master regulator of inflammatory responses. [1].[2].And MROCKI expression is association with violent suicide the mechanism of the MARCKS.[3].
Contents
Annotated Information
Name
Approved symbol:MROCKI
Approved name:MARCKS cis regulating lncRNA promoter of cytokines and inflammation
HGNC ID:50323
Previous names:long intergenic non-protein coding RNA 1268
Previous symbols:LINC01268
Alias symbols:LOC285758|ROCKI
RefSeq ID:NR_038863
Characteristics
MARCKS harbors an intragenic CpG island and is closely flanked by the uncharacterized MROCKI(hg19: chr6:114,189,179-114,194,512), which codes for an antisense transcript, a long noncoding RNA(ncRNA).[3]
Function
MROCKI changed expression in glioma is methylation-dependent and methylation correlates with WHO malignancy grade. Methylation is also distinctive between astrocytoma I-III and other glioma subtypes and may thus serve as an additional biomarker in glioma diagnosis.[1] MROCKI expression were linked to a reduced risk of certain inflammatory and infectious disease in humans, including inflammatory bowel disease and tuberculosis.[2] MROCKI is acted as a master regulator of inflammatory responses. [2] MROCKI expression is association with violent suicide the mechanism of the MARCKS.[3] Epigenetic changes associated with suicide represent two facets of epigenetic mechanisms related to MROCKI—that is, DNA methylation and ncRNA expression.[3]
Regulation
MROCKI interacted with APEX1 (apurinic/apyrimidinic endodeoxyribonuclease 1) to form a ribonucleoprotein complex at the MARCKS promoter. [2] MROCKI-APEX1 recruited the histone deacetylase HDAC1, which removed the H3K27ac modification from the promoter, thus reducing MARCKS transcription and subsequent Ca2+ signaling and inflammatory gene expression.[2]
Expression
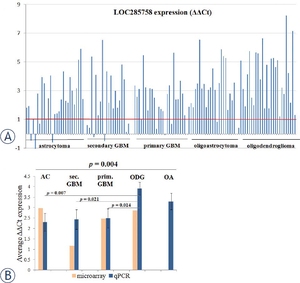
In all glioma subtypes levels of MROCKI were significantly higher in comparison to normal brain reference RNA, and expression was inversely associated with promoter methylation.[1] MROCKI expression is significantly increased in violent suicides irrespective of MARCKS transcription levels.[3]
Diseases
Glioma tumours[1]
Labs working on this lncRNA
- Lieber Institute for Brain Development, Johns Hopkins Medical Campus, Baltimore, MD, USA.[3]
- Group of Psychiatric Neuroscience, Department of Basic Medical Science, Neuroscience and Sense Organs, University of Bari Aldo Moro, Bari, Italy.[3]
- Section of Forensic Psychiatry and Criminology, Institute of Legal Medicine, University of Bari Aldo Moro, Bari, Italy.[3]
- Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD, USA.[3]
- Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD, USA.[3]
- Department of Molecular Genetics, Institute of Pathology, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia.[1]
- Institute of Pathology, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia.[1]
- Department of Neurosurgery, University Medical Center, Ljubljana, Slovenia.[1]
- Department of Pediatrics, University of California San Diego School of Medicine, La Jolla, CA, USA.[2]
- Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA.[2]
- Center for Infectious Disease Research (CIDR), Seattle, WA, USA.[2]
- Department of Immunology, University of Washington School of Medicine, Seattle, WA, USA.[2]
- Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego School of Medicine, La Jolla, CA, USA.[2]
- Department of Pediatrics, University of California San Diego School of Medicine, La Jolla, CA, USA trana@ucsd.edu.[2]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Matjasic A, Popovic M, Matos B, Glavac D. Expression of LOC285758, a Potential Long Non-coding Biomarker, is Methylation-dependent and Correlates with Glioma Malignancy Grade. Radiol Oncol. 2017 Jan 14;51(3):331-341. doi: 10.1515/raon-2017-0004.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 Zhang Q, Chao TC, Patil VS, Qin Y, Tiwari SK, Chiou J, Dobin A, Tsai CM, Li Z, Dang J, Gupta S, Urdahl K, Nizet V, Gingeras TR, Gaulton KJ, Rana TM. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019 Apr 15;38(8):e100041. doi: 10.15252/embj.2018100041. Epub 2019 Mar 27.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 3.9 Punzi G, Ursini G, Shin JH, Kleinman JE, Hyde TM, Weinberger DR. Increased expression of MARCKS in post-mortem brain of violent suicide completers is related to transcription of a long, noncoding, antisense RNA. Mol Psychiatry. 2014 Oct;19(10):1057-9. doi: 10.1038/mp.2014.41. Epub 2014 May 13.