Difference between revisions of "NONHSAT033797"
Chunlei Yu (talk | contribs) (→Function) |
|||
| (8 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | DLEU1 maps to a critical region at chromosomal band 13q14.3 which is recurrently deleted in solid tumors and hematopoietic malignancies like chronic lymphocytic leukemia (CLL). | |
==Annotated Information== | ==Annotated Information== | ||
| − | === | + | ===Name=== |
| + | [[File:Model of the tumor suppressor mechanism localized in 13q14.3.jpg|right|thumb|300px|'''Model of the tumor suppressor mechanism localized in 13q14.3'''<ref name="ref3" />.]] | ||
''DLEU1'':deleted in lymphocytic leukemia 1 (non-protein coding)(HGNC nomenclature) | ''DLEU1'':deleted in lymphocytic leukemia 1 (non-protein coding)(HGNC nomenclature) | ||
| Line 10: | Line 11: | ||
===Function=== | ===Function=== | ||
| − | + | DLEU1 or DLEU2 exert their function by binding to chromatin, but rather regulate the neighboring cluster of candidate tumor suppressor genes by divergent transcription <ref name="ref3" /> | |
| − | |||
| − | |||
===Regulation=== | ===Regulation=== | ||
| − | [[File:Downregulation of 13q14.3 candidate tumor suppressor genes and upregulation of lncRNA genes DLEU1 and DLEU2correlates with DNA-methylation.jpg|right|thumb| | + | [[File:Downregulation of 13q14.3 candidate tumor suppressor genes and upregulation of lncRNA genes DLEU1 and DLEU2correlates with DNA-methylation.jpg|right|thumb|300px|'''Downregulation of 13q14.3 candidate tumor suppressor genes and upregulation of lncRNA genes DLEU1 and DLEU2correlates with DNA-methylation.'''<ref name="ref3" />]] |
It was regulated by DNA-methylation and histone modifications<ref name="ref3" />. | It was regulated by DNA-methylation and histone modifications<ref name="ref3" />. | ||
| − | |||
| − | |||
===Expression=== | ===Expression=== | ||
| Line 46: | Line 43: | ||
tID = NONHSAT033797| | tID = NONHSAT033797| | ||
source = NONCODE4.0| | source = NONCODE4.0| | ||
| − | same = ''BCMS'', “deleted in lymphocytic leukemia, 1”, ''DLB1',"B-cell neoplasia-associated gene with multiple splicing", ''BCMS1'',LEU1, LINC00021, "long intergenic non-protein coding RNA 21", NCRNA00021, "non-protein coding RNA 21", XTP6| | + | same = ''DLEU1'',''BCMS'', “deleted in lymphocytic leukemia, 1”, ''DLB1',"B-cell neoplasia-associated gene with multiple splicing", ''BCMS1'',''LEU1'', ''LINC00021'', "long intergenic non-protein coding RNA 21", NCRNA00021, "non-protein coding RNA 21", ''XTP6''| |
classification = intergenic| | classification = intergenic| | ||
length = 1374 nt| | length = 1374 nt| | ||
Latest revision as of 12:47, 30 June 2016
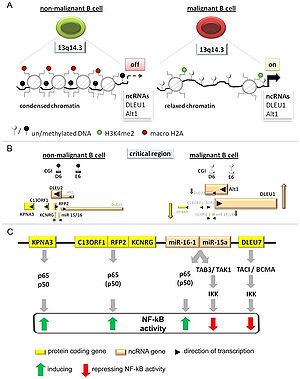
DLEU1 maps to a critical region at chromosomal band 13q14.3 which is recurrently deleted in solid tumors and hematopoietic malignancies like chronic lymphocytic leukemia (CLL).
Contents
Annotated Information
Name
DLEU1:deleted in lymphocytic leukemia 1 (non-protein coding)(HGNC nomenclature)
BCMS[2], “deleted in lymphocytic leukemia, 1”[2], DLB1[2],"B-cell neoplasia-associated gene with multiple splicing"[2], BCMS1[2],LEU1[3], LINC00021[3], "long intergenic non-protein coding RNA 21"[3], NCRNA00021[3], "non-protein coding RNA 21"[3], XTP6[3]
DLEU1: deleted in lymphocytic leukemia 2 (non-protein coding)(HGNC nomenclature)
Function
DLEU1 or DLEU2 exert their function by binding to chromatin, but rather regulate the neighboring cluster of candidate tumor suppressor genes by divergent transcription [1]
Regulation

It was regulated by DNA-methylation and histone modifications[1].
Expression
Both lncRNA genes DLEU1 and DLEU2/Alt1 were upregulated in Jurkat cells upon DNA-demethylation in-vitro , underlining that their transcriptional activity depends on levels of DNA-methylation.[1]
Disease
hematopoietic and solid tumors[1]
Labs working on this lncRNA
Abteilung Organisation Komplexer Genome (H0700), Deutsches Krebsforschungszentrum, Im Neuenheimer Feld 280, D-69120 Heidelberg, Germany.[2]
Radiumhemmet, Karolinska Hospital, Stockholm, Sweden.[3]
Cooperation Unit "Mechanisms of Leukemogenesis," University Ulm, German Cancer Research Center, DKFZ, Heidelberg, Germany.[1]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K, et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the In Cis downregulation of a gene cluster that targets NF-kB[J]. PLoS genetics. 2013,9(4):e1003373
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Wolf S, Mertens D, Schaffner C, Korz C, Dohner H, Stilgenbauer S, et al. B-cell neoplasia associated gene with multiple splicing (BCMS): the candidate B-CLL gene on 13q14 comprises more than 560 kb covering all critical regions[J]. Human molecular genetics. 2001,10(12):1275-85.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Liu Y, Corcoran M, Rasool O, Ivanova G, Ibbotson R, Grander D, et al. Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia[J]. Oncogene. 1997,15(20):2463-73.
Basic Information
| Transcript ID |
NONHSAT033797 |
| Source |
NONCODE4.0 |
| Same with |
DLEU1,BCMS, “deleted in lymphocytic leukemia, 1”, DLB1',"B-cell neoplasia-associated gene with multiple splicing", BCMS1,LEU1, LINC00021, "long intergenic non-protein coding RNA 21", NCRNA00021, "non-protein coding RNA 21", XTP6 |
| Classification |
intergenic |
| Length |
1374 nt |
| Genomic location |
chr13-:50618561..50679433 |
| Exon number |
6 |
| Exons |
50618561..50618906,50623720..50623800,50642233..50642301,50649695..50649789,50655959..50656147,50678840..50679433 |
| Genome context |
|
| Sequence |
000001 TTCATTCATC AAAGATTTAT TGAGTGCTTA CTGTGTGCCA GGCACTGTTC TAAAGACACA GCAGCAAACA AAACTGTTAT 000080
000081 AGAACAGGAT AAGGCAGGTC ATCTCCCAGA TACGACACTT TGATAGGAAC ACAAAATACA AATAGGAGAG TAGAAAGAGA 000160 000161 TGAAATACAA AGAGAAAGAG CTGATATTGG GCTTATATTA ATATTGAAAG GCCTTACATG CTAGATATTC TAAATGGGAG 000240 000241 AAACATGCTG GAAAGATACT TGGCATGAAT GAACTTATGT CCCTATGTTC AGAACAGACT GCTTTCCCTT CTCTGTTGGT 000320 000321 TGGCTCTTCT AGCCTCCTAG GAACTTTTAT AATCACCAGT ATGATCCAAT ATAAATACTG CACATATGAA GGTTCCCAGT 000400 000401 TGAGGAGTTT GGAAGCAAAA AGGGAAAGAA TGGCTGGCAA AGGCTTCAAT CTTCTGTCTC CTTTTATCTT CCCAATGGCT 000480 000481 AGTAGAAAAT ACTATCATCA ATCTCTTCCT TGCAACAATC TCCCATTTGA TATTTGTCTG CATTGTGACT CAATTCCTGA 000560 000561 AGGTTCTGTT CTTCATTCCT TTCTGGTAGA ATCAGGAGCC TCAGCGCTTT CGGCGCCATT TTCGAGTGAT GCCTGATCTC 000640 000641 ATCAATCTAG CGGGAGAGAC AGGATAACCT GTCCGAGAGT ATAGCGCCAC TATGACTCCG CCGGAAAAAT TACTTTAAAA 000720 000721 ATCGCCAAAA ATTACTTGGA GCAAAGGGCA GTCGGCGGAG CTTCGCCAAG GCTGGCGCAG TCGGTTTTGA CCTGTAGCAG 000800 000801 AGAACCAATT CTGGAGAACA GCCTCACTTC TTTGATTGAA TACTTACATA ATGCATTGGA ACATGACATG AGATTAAGAG 000880 000881 GTTTAATAAT GATAGAATGA AGACCACAAT AAAAGAGACC TCTACTTAGC TCAGCAATTC TTACCTTAGT CTTACCTATT 000960 000961 TGATGAAGAT GTCTTTTGAA AGGTGTACTG CAAGGAACAA AATGTTTGTA AATTCTCCTT TTACCAAGAG GTGGATAATT 001040 001041 ACTGTACCTT CCTCATGGAA AAAGTTTTAT TTAAAGTGTT ATTTCTCATT GAATACTATC AAAAAGGAAA AAAAAATGAC 001120 001121 CTAAACTTTT GAGATAGATT TGGCTCTAGT AAGTATTTAG TTATATCACT TGCATATCTG GGAGAAGAAA TAAGAGACTA 001200 001201 TCATCAGTAC ATTCCCATCT ACTAAAAAAA TTTATTTTAC ACATGTCAAG GGATTACTTA TAACTTCCAT TTTATTACTA 001280 001281 ATAGCTTGAA CCCTTTTAAT GAAGACCTAA CTCCTCCACC AGAAATTTAA GTTTATGTTC TTACTTTGTT TACTTATAAA 001360 001361 ATACATCTCA GGTA |