Difference between revisions of "ENST00000414769.1"
Chunlei Yu (talk | contribs) (→Transcriptomic Nomeclature) |
|||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
==Annotated Information== | ==Annotated Information== | ||
===Name=== | ===Name=== | ||
| − | MIR503HG: MIR503 host gene(HGNC | + | MIR503HG: MIR503 host gene [https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:28258 (HGNC:28258)] |
| − | + | ===Alias=== | |
| − | "H19 X-linked co-expressed lncRNA", ''H19X | + | "H19 X-linked co-expressed lncRNA", ''H19X'' <ref name="ref1" /> |
| + | ''MGC16121'' [https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:28258 (HGNC:28258)] | ||
===Characteristics=== | ===Characteristics=== | ||
| Line 18: | Line 19: | ||
Expression pattern appears to have dramatically shifted during evolution, from an ancestral testes-predominant pattern to preferential expression in the chorioallantoic placenta of eutherians<ref name="ref1" />. | Expression pattern appears to have dramatically shifted during evolution, from an ancestral testes-predominant pattern to preferential expression in the chorioallantoic placenta of eutherians<ref name="ref1" />. | ||
| + | ===Disease=== | ||
| + | Choriocarcinoma <ref name="ref1" /> | ||
===Evolution=== | ===Evolution=== | ||
| Line 24: | Line 27: | ||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
* Center for Integrative Genomics, University of Lausanne, 1015 Lausanne, Switzerland<ref name="ref1" />. | * Center for Integrative Genomics, University of Lausanne, 1015 Lausanne, Switzerland<ref name="ref1" />. | ||
| + | |||
| + | ==Sequence== | ||
| + | >gi|84848|ref|NR_024607.1| Homo sapiens MIR503 host gene (MIR503HG), long non-coding RNA | ||
| + | <dnaseq>GAAGGTAGAAGGTGGGGTCTGCCGGACGCGTGTTCCTGCCACCAGGTGCCCGCTCCCCGCGAGGCCGGCT | ||
| + | CAGGAGCAGAAGGAAGCCCGGTGCCAGCCAGCCTTCCTGAAAGACCAAGCCCGCGCCATCCGGCTTCCTC | ||
| + | CAGTGGACGCCTGCAGGACCCAGGAATGTTTTTCTTGAAGGCATCCAGCATCTCCAGTTAGCAGTACTGA | ||
| + | TTTTTTTTCCCCCCAACAAAGGAACACTACATCAACACTGTTGGCGGGGACCTGGACACAGAAGACTCCT | ||
| + | GTTTCAAGAAAATACAATCATCTCTCAAAGGCTGTAATTTATATGCATTTTAAAACTCTAGGCATTGAAA | ||
| + | ACCACCCAAGTGTCCCAAATAGAAGGGTAATATATAATCAATCACTCAGTGTAATATTATACATCCTTTA | ||
| + | AAAATGTTATTGGGAAATGTTTTATGATCTGTAAGTCCAAGGAATCCTCTCCCACCATTTCTTTCCCCCC | ||
| + | GCTGTTCCCCCATACCCACACTTCTTTGTTCCAATTGGCATGTAAATTTGGTTTTCCCGCCAAATGAGTC | ||
| + | AGTCATGATGGGAACCTCAACTGATTTGAACAGATGTGTGTCAATGTTACTTGGAAAACTAGATGTCAAT | ||
| + | AACCAGGGTCACAGAAAAAGGCAGTGGTCACAACTCTGTAAAAATGTATGCATGCACACAGACAAGAACT | ||
| + | AAAGTGGAACCCCACACAGGAAAACAGTGGGCTGTACTCCAGTGCTGGGACATTGAATGACTGTATGCTG | ||
| + | CTTTTGATTTCCGTTA</dnaseq> | ||
==References== | ==References== | ||
<references> | <references> | ||
| − | <ref name="ref1">Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods[J]. Nature. 2014,505(7485):635-40.</ref>(1) | + | <ref name="ref1"> Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods[J]. Nature. 2014,505(7485):635-40.</ref>(1) |
</references> | </references> | ||
Latest revision as of 02:31, 21 November 2018
MIR503HG, a lncRNA which is likely originated at least 370 Myr ago in the tetrapod ancestor, may functions like H19 by promoting miRNA transcription, preferentially in the placenta and in an imprinted manner.
Contents
Annotated Information
Name
MIR503HG: MIR503 host gene (HGNC:28258)
Alias
"H19 X-linked co-expressed lncRNA", H19X [1] MGC16121 (HGNC:28258)
Characteristics
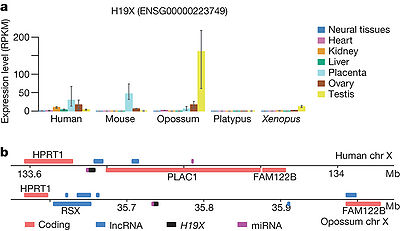
MIG503HG could potentially promote the transcription of between 2 and 7 miRNAs in different species, such as Human,mouse,Opossum,platyous,xenopus,. This lncRNA (that we name H19X, for H19 X-linked co-expressed lncRNA) is ikely originated at least 370 Myr ago, in the tetrapod ancestor[1].
Fuction
H19X is a neighbour of Rsx, the lncRNA that drives imprinted X-inactivation in marsupials, suggesting that H19X may itself be imprinted. These results suggest that H19X may function like H19, by promoting miRNA transcription, preferentially in the placenta and in an imprinted manner[1].
Expression
Expression pattern appears to have dramatically shifted during evolution, from an ancestral testes-predominant pattern to preferential expression in the chorioallantoic placenta of eutherians[1].
Disease
Choriocarcinoma [1]
Evolution
Expression pattern appears to have dramatically shifted during evolution, from an ancestral testes-predominant pattern to preferential expression in the chorioallantoic placenta of eutherians[1].
Labs working on this lncRNA
- Center for Integrative Genomics, University of Lausanne, 1015 Lausanne, Switzerland[1].
Sequence
>gi|84848|ref|NR_024607.1| Homo sapiens MIR503 host gene (MIR503HG), long non-coding RNA
000081 AGGAAGCCCG GTGCCAGCCA GCCTTCCTGA AAGACCAAGC CCGCGCCATC CGGCTTCCTC CAGTGGACGC CTGCAGGACC 000160
000161 CAGGAATGTT TTTCTTGAAG GCATCCAGCA TCTCCAGTTA GCAGTACTGA TTTTTTTTCC CCCCAACAAA GGAACACTAC 000240
000241 ATCAACACTG TTGGCGGGGA CCTGGACACA GAAGACTCCT GTTTCAAGAA AATACAATCA TCTCTCAAAG GCTGTAATTT 000320
000321 ATATGCATTT TAAAACTCTA GGCATTGAAA ACCACCCAAG TGTCCCAAAT AGAAGGGTAA TATATAATCA ATCACTCAGT 000400
000401 GTAATATTAT ACATCCTTTA AAAATGTTAT TGGGAAATGT TTTATGATCT GTAAGTCCAA GGAATCCTCT CCCACCATTT 000480
000481 CTTTCCCCCC GCTGTTCCCC CATACCCACA CTTCTTTGTT CCAATTGGCA TGTAAATTTG GTTTTCCCGC CAAATGAGTC 000560
000561 AGTCATGATG GGAACCTCAA CTGATTTGAA CAGATGTGTG TCAATGTTAC TTGGAAAACT AGATGTCAAT AACCAGGGTC 000640
000641 ACAGAAAAAG GCAGTGGTCA CAACTCTGTA AAAATGTATG CATGCACACA GACAAGAACT AAAGTGGAAC CCCACACAGG 000720
000721 AAAACAGTGG GCTGTACTCC AGTGCTGGGA CATTGAATGA CTGTATGCTG CTTTTGATTT CCGTTA
References
Basic Information
| Transcript ID |
ENST00000414769.1 |
| Source |
Gencode19 |
| Same with |
lnc-PLAC1-1:2, MIG403HG, H19X, MGC16121 |
| Classification |
intergenic |
| Length |
720 nt |
| Genomic location |
chrX-:133677473..133680660 |
| Exon number |
3 |
| Exons |
133677473..133678029,133678744..133678827,133680582..133680660 |
| Genome context |
|
| Sequence |
000001 GAAGGTAGAA GGTGGGGTCT GCCGGACGCG TGTTCCTGCC ACCAGGTGCC CGCTCCCCGC GAGGCCGGCT CAGGAGCAGA 000080
000081 AGGAAGCCCG GTGCCAGCCA GCCTTCCTGA AAGACCAAGC CCGCGCCATC CGGCTTCCTC CAGTGGACGC CTGCAGGACC 000160 000161 CAGGAATGTT TTTCTTGAAG GCATCCAGCA TCTCCAGTTA GCAGTACTGA TTTTTTTTCC CCCCAACAAA GGAACACTAC 000240 000241 ATCAACACTG TTGGCGGGGA CCTGGACACA GAAGACTCCT GTTTCAAGAA AATACAATCA TCTCTCAAAG GCTGTAATTT 000320 000321 ATATGCATTT TAAAACTCTA GGCATTGAAA ACCACCCAAG TGTCCCAAAT AGAAGGGTAA TATATAATCA ATCACTCAGT 000400 000401 GTAATATTAT ACATCCTTTA AAAATGTTAT TGGGAAATGT TTTATGATCT GTAAGTCCAA GGAATCCTCT CCCACCATTT 000480 000481 CTTTCCCCCC GCTGTTCCCC CATACCCACA CTTCTTTGTT CCAATTGGCA TGTAAATTTG GTTTTCCCGC CAAATGAGTC 000560 000561 AGTCATGATG GGAACCTCAA CTGATTTGAA CAGATGTGTG TCAATGTTAC TTGGAAAACT AGATGTCAAT AACCAGGGTC 000640 000641 ACAGAAAAAG GCAGTGGTCA CAACTCTGTA AAAATGTATG CATGCACACA GACAAGAACT AAAGTGGAAC CCCACACAGG 000720 |
Predicted Small Protein
| Name | ENST00000414769.1_smProtein_443:556 |
| Length | 38 |
| Molecular weight | 4124.8464 |
| Aromaticity | 0.135135135135 |
| Instability index | 65.8432432432 |
| Isoelectric point | 9.30364990234 |
| Runs | 5 |
| Runs residual | 0.0446317877511 |
| Runs probability | 0.0455043278572 |
| Amino acid sequence | MICKSKESSPTISFPPLFPHTHTSLFQLACKFGFPAK |
| Secondary structure | LEELLLLLLLLLLLLLLLLLLLHHHHHHHHHLLLLLL |
| PRMN | - |
| PiMo | - |
| Name | ENST00000414769.1_smProtein_443:556 |
| Length | 37 |
| Molecular weight | 4124.8464 |
| Aromaticity | 0.135135135135 |
| Instability index | 65.8432432432 |
| Isoelectric point | 9.30364990234 |
| Runs | 5 |
| Runs residual | 0.0446317877511 |
| Runs probability | 0.0455043278572 |
| Amino acid sequence | MICKSKESSPTISFPPLFPHTHTSLFQLACKFGFPAK |
| Secondary structure | LEELLLLLLLLLLLLLLLLLLLHHHHHHHHHLLLLLL |
| PRMN | - |
| PiMo | - |