Difference between revisions of "ENST00000608917.1"
Chunlei Yu (talk | contribs) (→Expression) |
|||
| Line 14: | Line 14: | ||
===Celluar Localization=== | ===Celluar Localization=== | ||
PACERR performs its functions in the nucleus<ref name="ref1" />. | PACERR performs its functions in the nucleus<ref name="ref1" />. | ||
| + | |||
| + | ===Disease=== | ||
| + | inflammation and cancer | ||
===Function=== | ===Function=== | ||
Latest revision as of 13:27, 1 July 2016
PACERR, a nuclear antisense long non-coding RNA PACER (P50-Associated COX-2 Extragenic RNA), which is expressed in the upstream region of COX-2 and functions to directly sequester the repressive NF-κB p50 subunit from the COX-2 promoter and activate the COX-2 gene, may function as an oncogene in osteosarcoma. [1][2]
Contents
Annotated Information
Name
PACERR,PTGS2 antisense NFKB1 complex-mediated expression regulator RNA(HGNC nomenclature)
"p50-associated COX-2 extragenic RNA", PACER, "PTGS2 antisense RNA 1 (head to head)", PTGS2-AS1[1]
Characteristics
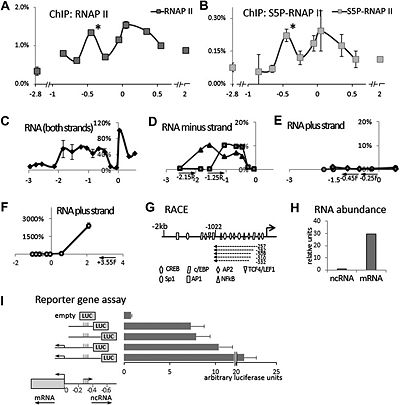
PACERR,homo sapiens PTGS2 antisense NFKB1 complex-mediated expression regulator RNA, was located in the upstream region of the human COX-2 gene[1].
Celluar Localization
PACERR performs its functions in the nucleus[1].
Disease
inflammation and cancer
Function

PACERR facilitates assembly of RNA Polymerase II pre-initiation complexes, through recruitment of the histone acetyltransferase p300 to catalyze histone acetylation at the COX-2 locus upon stimulus-induction[1].
PACERR directly interacts with the repressive NF-κB subunit p50 to occlude it from the COX-2 promoter[1].
PACERR belongs to a novel, unanticipated class of bona-fide transcriptional regulators[1].
The upregulation of PACER contributes to the dysregulation of COX-2 in osteosarcoma and subsequently promotes the development of osteosarcoma.[2]
Regulation
CTCF regulates the COX-2 locus by establishing and maintaining an open chromatin domain demarcated by two CTCF/cohesin complexes that is characterized by decreased H4K20 trimethylation, increased H3K4 di- and tri- methylation and increased histone acetylation[1].
Expression
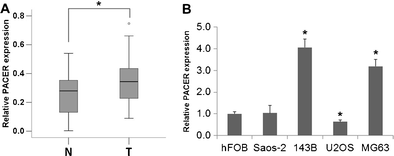
PACER was overexpressed in osteosarcoma tissues and cell lines compared with normal tissues and osteoblasts, respectively. [1]
PACER was overexpressed in osteosarcoma tissues and two osteosarcoma cell lines compared with normal tissues and osteoblasts, respectively.[2]
| Primers used in RACE | initial | nested |
| 5'RACE | TATGTATGTATGTGCTGCATATAG | ATGTCAGCCTTTCTTAACCTTAC[1] |
| 3'RACE | GTCTTTGCCCGAGCGCTTCCG | CCGCCGTGTCTGGTCTGTACGTC[1] |
| Experiment | Primer | Forward | Reverse |
| RNA quantitation | PACER | TGTAAATAGTTAATGTGAGCTCCACG | GCAAATTCTGGCCATCGC[1] |
| Experiment | Primer | Sequence |
| Knokdown | siPACER-753 | TGGAAAGAGAGGCGGGAAA[1] |
| siPACER-870 | TGGAAAGAGAGGCGGGAAA[1] |
Disease
PACER may be regarded as a therapeutic target in human osteosarcoma.[2]
Labs working on this lncRNA
- Regulatory Biology Laboratory, Salk Institute for Biological Studies, La Jolla, United States[1]
- Spine Tumor Center, Changzheng Hospital, Second Military Medical University, Fengyang Rd 415#, Huangpu District, Shanghai, China.[2]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes[J]. eLife. 2014,3:e01776.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Qian M, Yang X, Li Z, Jiang C, Song D, Yan W, et al. P50-associated COX-2 extragenic RNA (PACER) overexpression promotes proliferation and metastasis of osteosarcoma cells by activating COX-2 gene[J]. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016,37(3):3879-86.
Basic Information
| Transcript ID |
ENST00000608917.1 |
| Source |
Gencode19 |
| Same with |
PACER ,PTGS2-AS1 |
| Classification |
antisense |
| Length |
825 nt |
| Genomic location |
chr1+:186649754..186650578 |
| Exon number |
1 |
| Exons |
186649754..186650578 |
| Genome context |
|
| Sequence |
000001 AATTTGGGAG CAGAGGGGGT AGTCCCCACT CTCCTGTCTG ATCCCTCCCT CTCCTCCCCG AGTTCCACCG CCCCAGGCGC 000080
000081 ACAGGTTTCC GCCAGATGTC TTTTCTTCTT CGCAGTCTTT GCCCGAGCGC TTCCGAGAGC CAGTTCTGGA CTGATCGCCT 000160 000161 TGGATGGGAT ACCGGGGGAG GGCAGAAGGA CACTTGGCTT CCTCTCCAGG AATCTGAGCG GCCCTGAGGT CCGGGGGCGC 000240 000241 AGGGAATCCC CTCTCCCGCC GCCGCCGCCG TGTCTGGTCT GTACGTCTTT AGAGGGTCGA GGAAGTCACG TCGGGACAGA 000320 000321 CTGGGGCGAG TAAGGTTAAG AAAGGCTGAC ATGTTTTATG TTTTAGTGAC GACGCTTAAT AGGCTGTATA TCTGCTCTAT 000400 000401 ATGCAGCACA TACATACATA GCTTTTTAAA AAACTCTTAT TTTGTGGAAT GAAATAGCTA CCTTCAGTGT ACATAGCTGT 000480 000481 AATTTATCTT TGTAGCTAAG TTGCTTTCAA CAGAAGAAAT ACTGTTCTCC GTACCTTCAC CCCCTCCTTG TTTCTTGGAA 000560 000561 AGAGAGGCGG GAAAGGTAAA TTCTCCTCAT AATACTGGTC CTAAGCAGTT ACCCTGTAAA TAGTTAATGT GAGCTCCACG 000640 000641 GGTCACCAAT ATAAAGTTTC CTGCCTTCTG ATGGACAAAG GAAGCGGCGA TGGCCAGAAT TTGCAGGGAC GCTAAATGTC 000720 000721 CAAAACGTAT GCCTTAAGGC ATTTCTCTCC CTGATGCGTG GATTATTTTG GTTACTAGCC CTTCATAGGA GATACTGGTA 000800 000801 AAATAAATTC GAGTTTTAAA GTTCA |
Predicted Small Protein
| Name | ENST00000608917.1_smProtein_95:226 |
| Length | 44 |
| Molecular weight | 4914.4025 |
| Aromaticity | 0.116279069767 |
| Instability index | 79.0720930233 |
| Isoelectric point | 4.59381103516 |
| Runs | 5 |
| Runs residual | 0.0410243010191 |
| Runs probability | 0.0137078254725 |
| Amino acid sequence | MSFLLRSLCPSASESQFWTDRLGWDTGGGQKDTWLPLQESERP |
| Secondary structure | LLEEELLLLLLLLLLLLHHHHLLLLLLLLLLLLLLLLLLLLLL |
| PRMN | - |
| PiMo | - |