Difference between revisions of "PRAL"
Jiabao Cao (talk | contribs) (→Function) |
|||
| (10 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | ''PRAL'', p53 regulation associated lncRNA is a long non-coding RNAs (lncRNAs) play a crucial role in pathogenesis of diverse types of cancers. | ||
==Annotated Information== | ==Annotated Information== | ||
===Name=== | ===Name=== | ||
| − | PRAL:p53 regulation associated lncRNA | + | ''PRAL'': p53 regulation associated lncRNA (HGNC nomenclature), LncRNA-PRAL <ref name="ref1" />. |
| − | |||
===Characteristics=== | ===Characteristics=== | ||
| − | LncRNA-PRAL is transcribed with a poly | + | LncRNA-PRAL localizes on chromosome 17p13.1, and is transcribed with a poly A tail <ref name="ref1" />. |
| − | + | Most of the sequence of the lncRNA-PRAL overlaps in antisense with the 3' untranslated region of XAF1 protein-coding gene, which is known to regulate apoptosis<ref name="ref1" />. | |
===Cellular Localization=== | ===Cellular Localization=== | ||
| − | LncRNA-PRAL | + | LncRNA-PRAL is diffusely distributed both in the nucleus and cytoplasm, and localizes to both cytoplasmic and nuclear regions <ref name="ref1" />. |
| − | === | + | ===Function=== |
| + | [[File: PLAR.jpg|right|thumb|400px|'''Biological effects of long noncoding RNA ''PRAL'' in hepatocellular carcinoma (HCC).''' <ref name="ref1" />.]] | ||
| + | Somatic copy number variation of lncRNA-PRAL is associated with hepatocellular carcinoma (HCC) growth and lncRNA-PRAL may serve as a potential target for antitumor therapy <ref name="ref1" />. LncRNA-PRAL could inhibit HCC growth and induce apoptosis in vivo and in vitro through p53 <ref name="ref1" />. The three stem-loop motifs at the 5' end of lncRNA-PRAL facilitates the combination of HSP90 and p53 and thus competitively inhibites MDM2-dependent p53 ubiquitination, resulting in enhanced p53 stability <ref name="ref1" />. | ||
| + | ''PRAL'' might be a tumor suppressor in lung cancer <ref name="ref2" />. The transcript level of PRAL is decreased in lung cancer in vivo and in vitro, and overexpression of ''PRAL'' inhibits cell proliferation by upregulating the expression of P53 <ref name="ref2" />. | ||
| − | + | ''PRAL'' promotes multiple myeloma (MM) cell growth inhibition and apoptosis, and improves the bortezomib (BTZ) sensitivity in vitro <ref name="ref3" />. | |
| − | + | ||
| − | + | ===Expression=== | |
| + | Expression level of ''PRAL'' is decreased in lung cancer <ref name="ref2" />. | ||
| − | + | ''PRAL'' is downregulated in primary multiple myeloma (MM) cells and cell lines <ref name="ref3" />. | |
| + | {| class='wikitable' style="text-align:center" | ||
| + | |- | ||
| + | ! | Experiment | ||
| + | ! | Forward primer | ||
| + | ! | Reverse primer | ||
| + | |- | ||
| + | | rowspan="1"|Real Time PCR | ||
| + | | | 5′-GGCAGAGTCTCGCTTGGT-3′ | ||
| + | | | 5′-GAAACTCC GTCTCCGCTAA-3′<ref name="ref2" /> | ||
| + | |} | ||
===Diseases=== | ===Diseases=== | ||
| − | *Hepatocellular Carcinoma<ref name="ref1" /> | + | * Hepatocellular Carcinoma <ref name="ref1" /> |
| − | *Lung Cancer<ref name="ref2" /> | + | * Lung Cancer <ref name="ref2" /> |
| − | + | * Multiple myeloma (MM) <ref name="ref3" /> | |
| + | * Non-small-cell lung cancer (NSCLC) <ref name="ref4" /> | ||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
| − | *Department of Medical Genetics, Second Military Medical University, Shanghai, 200433, China. | + | * Department of Medical Genetics, Second Military Medical University, Shanghai, 200433, China.<ref name="ref1" /> |
| − | *The Third Department of Hepatic Surgery, Eastern Hepatobiliary Hospital, Second Military Medical University, Shanghai, 200433, China. | + | * The Third Department of Hepatic Surgery, Eastern Hepatobiliary Hospital, Second Military Medical University, Shanghai, 200433, China.<ref name="ref1" /> |
| − | *Department of Epidemiology, Second Military Medical University, Shanghai, 200433, China.<ref name="ref1" /> | + | * Department of Epidemiology, Second Military Medical University, Shanghai, 200433, China.<ref name="ref1" /> |
| − | *Department of Surgery, Xi'an Red Cross Hospital, Affiliated to School of Medicine, Xi'an Jiao Tong University, Xi'an, Shanxi, China(mainland). | + | * Department of Surgery, Xi'an Red Cross Hospital, Affiliated to School of Medicine, Xi'an Jiao Tong University, Xi'an, Shanxi, China(mainland).<ref name="ref2" /> |
| − | *Maternal and Child Care Service Center of Changan District, Xi'an, Shanxi, China (mainland)<ref name="ref2" />. | + | * Maternal and Child Care Service Center of Changan District, Xi'an, Shanxi, China (mainland).<ref name="ref2" /> |
| − | + | * Hematology of Shanghai Xuhui Centre Hospital, Shanghai Clinical Centre of Chinese Acadymy of Sciences, CAS, Shanghai 200031, PR China.<ref name="ref3" /> | |
==References== | ==References== | ||
<references> | <references> | ||
| − | <ref name="ref1"> | + | <ref name="ref1"> Zhou CC, Yang F, Yuan SX, Ma JZ, Liu F, Yuan JH, Bi FR, Lin KY, Yin JH, Cao GW, Zhou WP, Wang F, Sun SH. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology. 2016 Mar;63(3):850-63. </ref> |
| − | Zhou CC, Yang F, Yuan SX, Ma JZ, Liu F, Yuan JH, Bi FR, Lin KY, Yin JH, Cao | + | <ref name="ref2"> Su P, Wang F, Qi B, Wang T, Zhang S. P53 Regulation-Association Long Non-Coding RNA (LncRNA PRAL) Inhibits Cell Proliferation by Regulation of P53 in Human Lung Cancer. Med Sci Monit. 2017 Apr 11;23:1751-1758.</ref> |
| − | GW, Zhou WP, Wang F, Sun SH. Systemic genome screening identifies the outcome | + | <ref name="ref3"> Xiao G, Li Y, Wang Y, Zhao B, Zou Z, Hou S et al. LncRNA PRAL is closely related to clinical prognosis of multiple myeloma and the bortezomib sensitivity[J]. Experimental Cell Research. 2018.</ref> |
| − | associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. | + | <ref name="ref4"> Wang H, Wang J, Liang CF, Wang & Zhou T. Expression of Long Non-Coding RNA PRAL as a Potential Biomarker for Diagnosis in Non-Small-Cell Lung Cancer Patients is Associated with the Inhibition of Cell Proliferation and Metastasis[J]. Clinical laboratory. 2018, 64(9):1341-1348.</ref> |
| − | Hepatology. 2016 Mar;63(3):850-63. | ||
| − | </ref> | ||
| − | <ref name="ref2"> | ||
| − | Su P, Wang F, Qi B, Wang T, Zhang S. P53 Regulation-Association Long | ||
| − | Non-Coding RNA (LncRNA PRAL) Inhibits Cell Proliferation by Regulation of P53 in | ||
| − | Human Lung Cancer. Med Sci Monit. 2017 Apr 11;23:1751-1758. | ||
| − | </ref> | ||
</references> | </references> | ||
| + | |||
| + | ===Sequence=== | ||
| + | >ENST00000624952.1| Homo sapiens p53 regulation associated lncRNA (PRAL), long non-coding RNA | ||
| + | <dnaseq> | ||
| + | AAACCTGCAGTTGCCTGGTCAGTGGATCCCATGTGCAGAGGGCGAGACGGGGCCCTGGCC | ||
| + | AGGCGTTGGGAGGCGGGAGGCTCGGCCGGCGCTCCGGGTGAGGATGGCACGCCCCCGCCC | ||
| + | TTGCTTCTGCGCACAGGGAAGACATTCGGCCCCGAACTTGGGGCTCAAAACGGTTCAGTT | ||
| + | CGCTGCCTTTCCCATCCTGACCCAGCCCCGGGTTTTTTCCGAAGCCGCCGCTGGATAGGT | ||
| + | TGCATTACTAGGCTTCCCCCAGCCACTCTCCCTGCCCAAAGCCTTCAGAAGCCGCCCCTC | ||
| + | TCCCCTTCACCTGGCCCTTATCCACTGTCCCAGCCTCATTTCTTTCCCCCTCCCTGAATG | ||
| + | TCTTCTGCTCCCCTTTCCTTGGGCACTGCCCTTATCCTGGGGTCCTGGCTGCTGCCATTC | ||
| + | CTGGCCCTGGAAAACCAGTGCGGAACCCAAAGAAAGGTGGAAGAGCAGCTTTTAGAGAGG | ||
| + | TGTTTTTATTCTTTGGGGACAAAGCCGGGTTCTGTGGGTGTAGGATTCTCCAGGTTCTCC | ||
| + | AGGCTGTAGGGCCCAGAGGCTTAATCAGAATTTTCAGACAAAACTGGAACCTTTCTTTTT | ||
| + | TCCCGTTGGTTTATTTGTAGTCCTTGGGCAAACCAATGTCTTTGTTCGAAAGAGGGAAAA | ||
| + | TAATCCAAACGTTTTTCTTTTAACTTTTTTTTTAGGTTCAGGGGCACATGTGTAGGCTTG | ||
| + | CTATATAGGTAAATTGCATGTCACCAGGGTTTGTTGTACAGATTATTTCATCATCCAGAT | ||
| + | AAAAAGCATAGTACCAGATAGGTAGTTTTTTGATCCTCACCCTCCTTCCATGCTCCGACC | ||
| + | TCAGGTAGGCCCCAGTGTCTGCGGTTCCCTTCTTTGTATCCATGTGTACTCAATGTTTAG | ||
| + | CTTCTACTTATAAGTGAGAACATGTGGTATTTGGTTTTCTGTTTCTGAGTGAGTTCATTT | ||
| + | AGGATAATGGCTTCCAGCTCCATCCACGTTGCTGCACAGGACATGATCCCATTCTTTTTT | ||
| + | ATGGCTGCATAGTATTCCATGGTGTATATGTGCCACATTTTCTTTTTTTTTTTTTTAAGG | ||
| + | CAGAGTCTCGCTTGGTTGCCCAGGCTGGAGTGCAGTGGCAGGATCTCGGCTCACTGCAAC | ||
| + | CACCACCTTCCAGGTTCAAGCGATTCTCACGCCTAGGCCTCCCAAGTAGCTGGGATGACA | ||
| + | GGTGCTCGCCACCACGCCTGGCTAATTTTCGTATTTTTAGCGGAGACGGAGTTTCTCCAT | ||
| + | GTTGGCCAGGCTGGTCTCAAACTCCTGACCTCAGGTGAACCACCCGCCTCAGCCTCCCAA | ||
| + | AGTGCTGGGATTACAGACATGAGCCACCACTCCCTGCCACCACATTTTCTTTATCCTTTC | ||
| + | TACCACTGATGGACATTTAGTTTGATTCCCTGACTTTGCGATTGTGTATAGTGCTGCCGT | ||
| + | GAACATTTGTGTATATATGTCTTTATGACAGAATGATTTAGATTCCTTTGGGTATATAAC | ||
| + | CAATAATGGGATTGCTGAGTCAAATGGTAATTCTGTTTAATCATGTCCCATTTGTCAATT | ||
| + | TTTGCTTTCGTTACAATTATTTTTGGGATCTTTGTCATGAAATCTTTTTCCGTTCCTATG | ||
| + | TCCAGAATGGTATTGCCTAGGTTATCTTCCAGAGTTTTTATAGTTTGGGGTTTTACATTT | ||
| + | AAGTCTTTAATCCATCTTGAGTTGATTTTTGCATATGGTGTAAGGAAGTGGTTCAGTTTC | ||
| + | AATCCTCTGCATATGGCTAGCTAGTTATCCCAGTACCACTTACTGAATGGGGAATTCTTT | ||
| + | CCCCATTGCGTATTTTTGTCAACTTTGTCAAAGATTAGATGGTTGTAGGTGTGTAGCTTT | ||
| + | ATTTCTGGGCCCTCTATTCTGTTCCATTGATCTATGTGTCTGCTTTTGTACCAGCACCAT | ||
| + | GCTTTTGTACCAGTATCATGCAGTTTTGGTTACTGTAGCCCTGTAGTATAGTTTGAAGTT | ||
| + | GGGTAATGCGATGCCTCCAGCTTTGTTCTTTTTGCTTAGGATTGCTTTGGCTTTTTGGGC | ||
| + | TCTTTTTTGGTTTCAAATAAATTTTAATGCTTTTTTTTTCTGATTCTGTGAAGAATGTTA | ||
| + | TTGGTAGTTTGATAGGAATAGCATTGAATCTATAAATTGCTTTGGGTAGTATGGCCATTT | ||
| + | TAACAATATTGATTCTTCCTATCCATAAGTATGGAATGTTTTTCCATTTGTGTCATCTCT | ||
| + | GATTTCTTTCAGCAGTGTTTTGTAATTTTTGTTGTAGAGATCTTTCATCTCCCTGGTTAG | ||
| + | CTGTATTCCTAGGTATTTTATTCTTTTTGTGGCAATTGTGAATAGGATTGCATTCTTGAT | ||
| + | TTGGCTCTCAGCTTGGATGTCGTTGGTGTATAAAAATGCTACCATATTTTGTATATTGAT | ||
| + | TTTGTCTCCTGAAAGTTAGCTGAAGTTGTTTATCAGATCTAGGAGCTTTTGGGCCAAGAT | ||
| + | CATGGGGTTTTCTAGATATAGAATCATATCGTCTGCAAACAGAGATACTTCGACTTCCTT | ||
| + | TCTTTTTGGTTGCCTTTTATTTCTTTCTCTTGCCTGATTGCTGTGGCGAGGACTTCCAGT | ||
| + | ACTATGTTGAATAGGACTGAAGTGCCTTGTTCTACTTCTCAAGAGTAATGCTCCAGCTTT | ||
| + | TGCTCATTCAGTATGATGTTGGCTGTGGTTTTGTCATAGATGGCTCTTATTATTTTGAGG | ||
| + | TATGTTCCTTCGACGCCTGGTTTGTTGAGGGTTTTTAACATGAAGTCAAGTTGAATTTTA | ||
| + | TCAAAAGCCTTTTCTGTATCTATTGAGATGATACGTGAGTTTTGTTTTTAGTTCTGCCAA | ||
| + | ACCTTTTCTTTTAAAGCCCAACGAATAAAACCCATCACCTTTCACAAGACCACCACAGCA | ||
| + | AGTAGGCAGGAATGCCAGTGTTAAAAGTGAAATCTTTTGAATTTGTAGTACCTTTCCTTT | ||
| + | TCCAAATCTAGCTGAAATTTCTCACTTGTTTTCCTTTTGATGAAGCTAACCACCGGCATT | ||
| + | TCTCCTAAGAAATGGATATAAAAAAAGTTTGATATGGTTAAAGTAAGAAGTGCTAGCTCC | ||
| + | TAGAATATCTCTGGATATTGCTGTCCCTTGTTACAGAGCATGAGCCCTGCCCAAAGACTG | ||
| + | GAGTGTAATGGCATAGAAAAGGTCAGGAGCCCACTGAGCCAGAAGTATAGGAGGCTTGCG | ||
| + | ACAAAGTTTAACAGGGGCCTGTTGCTATCACTGGGTGGAGGGACAGATGGGAGAACACTC | ||
| + | ACACCATGGAATCTTAGGCAGCCATTAATAAAAATAAGCCACACTG</dnaseq> | ||
Latest revision as of 03:27, 26 November 2018
PRAL, p53 regulation associated lncRNA is a long non-coding RNAs (lncRNAs) play a crucial role in pathogenesis of diverse types of cancers.
Contents
Annotated Information
Name
PRAL: p53 regulation associated lncRNA (HGNC nomenclature), LncRNA-PRAL [1].
Characteristics
LncRNA-PRAL localizes on chromosome 17p13.1, and is transcribed with a poly A tail [1]. Most of the sequence of the lncRNA-PRAL overlaps in antisense with the 3' untranslated region of XAF1 protein-coding gene, which is known to regulate apoptosis[1].
Cellular Localization
LncRNA-PRAL is diffusely distributed both in the nucleus and cytoplasm, and localizes to both cytoplasmic and nuclear regions [1].
Function
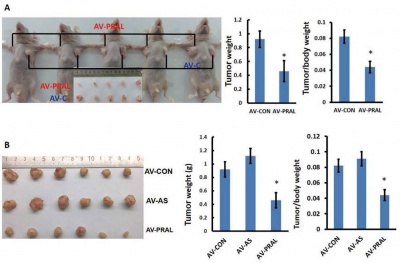
Somatic copy number variation of lncRNA-PRAL is associated with hepatocellular carcinoma (HCC) growth and lncRNA-PRAL may serve as a potential target for antitumor therapy [1]. LncRNA-PRAL could inhibit HCC growth and induce apoptosis in vivo and in vitro through p53 [1]. The three stem-loop motifs at the 5' end of lncRNA-PRAL facilitates the combination of HSP90 and p53 and thus competitively inhibites MDM2-dependent p53 ubiquitination, resulting in enhanced p53 stability [1].
PRAL might be a tumor suppressor in lung cancer [2]. The transcript level of PRAL is decreased in lung cancer in vivo and in vitro, and overexpression of PRAL inhibits cell proliferation by upregulating the expression of P53 [2].
PRAL promotes multiple myeloma (MM) cell growth inhibition and apoptosis, and improves the bortezomib (BTZ) sensitivity in vitro [3].
Expression
Expression level of PRAL is decreased in lung cancer [2].
PRAL is downregulated in primary multiple myeloma (MM) cells and cell lines [3].
| Experiment | Forward primer | Reverse primer |
|---|---|---|
| Real Time PCR | 5′-GGCAGAGTCTCGCTTGGT-3′ | 5′-GAAACTCC GTCTCCGCTAA-3′[2] |
Diseases
- Hepatocellular Carcinoma [1]
- Lung Cancer [2]
- Multiple myeloma (MM) [3]
- Non-small-cell lung cancer (NSCLC) [4]
Labs working on this lncRNA
- Department of Medical Genetics, Second Military Medical University, Shanghai, 200433, China.[1]
- The Third Department of Hepatic Surgery, Eastern Hepatobiliary Hospital, Second Military Medical University, Shanghai, 200433, China.[1]
- Department of Epidemiology, Second Military Medical University, Shanghai, 200433, China.[1]
- Department of Surgery, Xi'an Red Cross Hospital, Affiliated to School of Medicine, Xi'an Jiao Tong University, Xi'an, Shanxi, China(mainland).[2]
- Maternal and Child Care Service Center of Changan District, Xi'an, Shanxi, China (mainland).[2]
- Hematology of Shanghai Xuhui Centre Hospital, Shanghai Clinical Centre of Chinese Acadymy of Sciences, CAS, Shanghai 200031, PR China.[3]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Zhou CC, Yang F, Yuan SX, Ma JZ, Liu F, Yuan JH, Bi FR, Lin KY, Yin JH, Cao GW, Zhou WP, Wang F, Sun SH. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology. 2016 Mar;63(3):850-63.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Su P, Wang F, Qi B, Wang T, Zhang S. P53 Regulation-Association Long Non-Coding RNA (LncRNA PRAL) Inhibits Cell Proliferation by Regulation of P53 in Human Lung Cancer. Med Sci Monit. 2017 Apr 11;23:1751-1758.
- ↑ 3.0 3.1 3.2 3.3 Xiao G, Li Y, Wang Y, Zhao B, Zou Z, Hou S et al. LncRNA PRAL is closely related to clinical prognosis of multiple myeloma and the bortezomib sensitivity[J]. Experimental Cell Research. 2018.
- ↑ Wang H, Wang J, Liang CF, Wang & Zhou T. Expression of Long Non-Coding RNA PRAL as a Potential Biomarker for Diagnosis in Non-Small-Cell Lung Cancer Patients is Associated with the Inhibition of Cell Proliferation and Metastasis[J]. Clinical laboratory. 2018, 64(9):1341-1348.
Sequence
>ENST00000624952.1| Homo sapiens p53 regulation associated lncRNA (PRAL), long non-coding RNA
000081 CTCGGCCGGC GCTCCGGGTG AGGATGGCAC GCCCCCGCCC TTGCTTCTGC GCACAGGGAA GACATTCGGC CCCGAACTTG 000160
000161 GGGCTCAAAA CGGTTCAGTT CGCTGCCTTT CCCATCCTGA CCCAGCCCCG GGTTTTTTCC GAAGCCGCCG CTGGATAGGT 000240
000241 TGCATTACTA GGCTTCCCCC AGCCACTCTC CCTGCCCAAA GCCTTCAGAA GCCGCCCCTC TCCCCTTCAC CTGGCCCTTA 000320
000321 TCCACTGTCC CAGCCTCATT TCTTTCCCCC TCCCTGAATG TCTTCTGCTC CCCTTTCCTT GGGCACTGCC CTTATCCTGG 000400
000401 GGTCCTGGCT GCTGCCATTC CTGGCCCTGG AAAACCAGTG CGGAACCCAA AGAAAGGTGG AAGAGCAGCT TTTAGAGAGG 000480
000481 TGTTTTTATT CTTTGGGGAC AAAGCCGGGT TCTGTGGGTG TAGGATTCTC CAGGTTCTCC AGGCTGTAGG GCCCAGAGGC 000560
000561 TTAATCAGAA TTTTCAGACA AAACTGGAAC CTTTCTTTTT TCCCGTTGGT TTATTTGTAG TCCTTGGGCA AACCAATGTC 000640
000641 TTTGTTCGAA AGAGGGAAAA TAATCCAAAC GTTTTTCTTT TAACTTTTTT TTTAGGTTCA GGGGCACATG TGTAGGCTTG 000720
000721 CTATATAGGT AAATTGCATG TCACCAGGGT TTGTTGTACA GATTATTTCA TCATCCAGAT AAAAAGCATA GTACCAGATA 000800
000801 GGTAGTTTTT TGATCCTCAC CCTCCTTCCA TGCTCCGACC TCAGGTAGGC CCCAGTGTCT GCGGTTCCCT TCTTTGTATC 000880
000881 CATGTGTACT CAATGTTTAG CTTCTACTTA TAAGTGAGAA CATGTGGTAT TTGGTTTTCT GTTTCTGAGT GAGTTCATTT 000960
000961 AGGATAATGG CTTCCAGCTC CATCCACGTT GCTGCACAGG ACATGATCCC ATTCTTTTTT ATGGCTGCAT AGTATTCCAT 001040
001041 GGTGTATATG TGCCACATTT TCTTTTTTTT TTTTTTAAGG CAGAGTCTCG CTTGGTTGCC CAGGCTGGAG TGCAGTGGCA 001120
001121 GGATCTCGGC TCACTGCAAC CACCACCTTC CAGGTTCAAG CGATTCTCAC GCCTAGGCCT CCCAAGTAGC TGGGATGACA 001200
001201 GGTGCTCGCC ACCACGCCTG GCTAATTTTC GTATTTTTAG CGGAGACGGA GTTTCTCCAT GTTGGCCAGG CTGGTCTCAA 001280
001281 ACTCCTGACC TCAGGTGAAC CACCCGCCTC AGCCTCCCAA AGTGCTGGGA TTACAGACAT GAGCCACCAC TCCCTGCCAC 001360
001361 CACATTTTCT TTATCCTTTC TACCACTGAT GGACATTTAG TTTGATTCCC TGACTTTGCG ATTGTGTATA GTGCTGCCGT 001440
001441 GAACATTTGT GTATATATGT CTTTATGACA GAATGATTTA GATTCCTTTG GGTATATAAC CAATAATGGG ATTGCTGAGT 001520
001521 CAAATGGTAA TTCTGTTTAA TCATGTCCCA TTTGTCAATT TTTGCTTTCG TTACAATTAT TTTTGGGATC TTTGTCATGA 001600
001601 AATCTTTTTC CGTTCCTATG TCCAGAATGG TATTGCCTAG GTTATCTTCC AGAGTTTTTA TAGTTTGGGG TTTTACATTT 001680
001681 AAGTCTTTAA TCCATCTTGA GTTGATTTTT GCATATGGTG TAAGGAAGTG GTTCAGTTTC AATCCTCTGC ATATGGCTAG 001760
001761 CTAGTTATCC CAGTACCACT TACTGAATGG GGAATTCTTT CCCCATTGCG TATTTTTGTC AACTTTGTCA AAGATTAGAT 001840
001841 GGTTGTAGGT GTGTAGCTTT ATTTCTGGGC CCTCTATTCT GTTCCATTGA TCTATGTGTC TGCTTTTGTA CCAGCACCAT 001920
001921 GCTTTTGTAC CAGTATCATG CAGTTTTGGT TACTGTAGCC CTGTAGTATA GTTTGAAGTT GGGTAATGCG ATGCCTCCAG 002000
002001 CTTTGTTCTT TTTGCTTAGG ATTGCTTTGG CTTTTTGGGC TCTTTTTTGG TTTCAAATAA ATTTTAATGC TTTTTTTTTC 002080
002081 TGATTCTGTG AAGAATGTTA TTGGTAGTTT GATAGGAATA GCATTGAATC TATAAATTGC TTTGGGTAGT ATGGCCATTT 002160
002161 TAACAATATT GATTCTTCCT ATCCATAAGT ATGGAATGTT TTTCCATTTG TGTCATCTCT GATTTCTTTC AGCAGTGTTT 002240
002241 TGTAATTTTT GTTGTAGAGA TCTTTCATCT CCCTGGTTAG CTGTATTCCT AGGTATTTTA TTCTTTTTGT GGCAATTGTG 002320
002321 AATAGGATTG CATTCTTGAT TTGGCTCTCA GCTTGGATGT CGTTGGTGTA TAAAAATGCT ACCATATTTT GTATATTGAT 002400
002401 TTTGTCTCCT GAAAGTTAGC TGAAGTTGTT TATCAGATCT AGGAGCTTTT GGGCCAAGAT CATGGGGTTT TCTAGATATA 002480
002481 GAATCATATC GTCTGCAAAC AGAGATACTT CGACTTCCTT TCTTTTTGGT TGCCTTTTAT TTCTTTCTCT TGCCTGATTG 002560
002561 CTGTGGCGAG GACTTCCAGT ACTATGTTGA ATAGGACTGA AGTGCCTTGT TCTACTTCTC AAGAGTAATG CTCCAGCTTT 002640
002641 TGCTCATTCA GTATGATGTT GGCTGTGGTT TTGTCATAGA TGGCTCTTAT TATTTTGAGG TATGTTCCTT CGACGCCTGG 002720
002721 TTTGTTGAGG GTTTTTAACA TGAAGTCAAG TTGAATTTTA TCAAAAGCCT TTTCTGTATC TATTGAGATG ATACGTGAGT 002800
002801 TTTGTTTTTA GTTCTGCCAA ACCTTTTCTT TTAAAGCCCA ACGAATAAAA CCCATCACCT TTCACAAGAC CACCACAGCA 002880
002881 AGTAGGCAGG AATGCCAGTG TTAAAAGTGA AATCTTTTGA ATTTGTAGTA CCTTTCCTTT TCCAAATCTA GCTGAAATTT 002960
002961 CTCACTTGTT TTCCTTTTGA TGAAGCTAAC CACCGGCATT TCTCCTAAGA AATGGATATA AAAAAAGTTT GATATGGTTA 003040
003041 AAGTAAGAAG TGCTAGCTCC TAGAATATCT CTGGATATTG CTGTCCCTTG TTACAGAGCA TGAGCCCTGC CCAAAGACTG 003120
003121 GAGTGTAATG GCATAGAAAA GGTCAGGAGC CCACTGAGCC AGAAGTATAG GAGGCTTGCG ACAAAGTTTA ACAGGGGCCT 003200
003201 GTTGCTATCA CTGGGTGGAG GGACAGATGG GAGAACACTC ACACCATGGA ATCTTAGGCA GCCATTAATA AAAATAAGCC 003280
003281 ACACTG