Difference between revisions of "SMILR"
| (12 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | ''SMILR'' (smooth muscle induced lncRNA, enhancer of proliferationas) is a long noncoding RNA that is used as a driver of vascular smooth muscle cell proliferation and consider as a novel therapeutic strategy to reduce vascular pathologies. | ||
==Annotated Information== | ==Annotated Information== | ||
| − | === | + | ===Name=== |
| − | SMILR: | + | ''SMILR'': Smooth muscle induced lncRNA, enhancer of proliferation (HGNC nomenclature), RP11-94a24.1 <ref name="ref1" /> |
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
===Characteristics=== | ===Characteristics=== | ||
| − | + | [[File:Localization of SMILR.jpg|right|thumb|300px|'''Localization of SMILR.(A)RNA FISH analysis of ''SMILR'',cytoplasmic UBC mRNA,and nuclear SNORD3 RNA in HSVSMC.(B)25±5 individual SMILR molecules were observed within the nucleus and cytoplasm.''' <ref name="ref1" />.]] | |
| − | [[File:Localization of SMILR.jpg|right|thumb|300px|'''Localization of SMILR.(A)RNA FISH analysis of SMILR,cytoplasmic UBC mRNA,and nuclear SNORD3 RNA in HSVSMC.(B)25±5 individual SMILR molecules were observed within the nucleus and cytoplasm.''' <ref name="ref1" />.]] | + | ''SMILR'' is four exon transcript of 569 bp long noncoding RNA that is located on human chromosome 8q24.13, ~750 kbp downstream of ''HAS2'' on the same (reverse) strand and ~350 kbp from ''ZHX2'' and ~750 kbp from ''HAS2-AS1'' on the opposite strand of chromosome 8 <ref name="ref1" />. The longest open reading frame(ORF) within SMILR is 57 amino acids <ref name="ref1" />. |
| − | === | + | ''SMILR'' is located in both nucleus and cytoplasm <ref name="ref1" />. |
| − | SMILR | + | |
| + | ===Function=== | ||
| + | [[File:SMILR.jpg|right|thumb|300px|'''''SMILR'' regulates proximal gene HAS2 in chromosome 8.''' <ref name="ref1" />.]] | ||
| + | ''SMILR'' acts as a driver of vascular smooth muscle cell proliferation <ref name="ref1" />. Knockdown of ''SMILR'' markedly reduced cell proliferation and HAS2 expression was also reduced by ''SMILR'' knockdown <ref name="ref1" />. | ||
===Expression=== | ===Expression=== | ||
| − | SMILR is expressed highest in heart <ref name="ref1" />. It is differentially expressed in human saphenous vein-derived smooth muscle cells following stimulation with interleukin-1α and platelet-derived growth factor <ref name="ref1" />. Increased expression of SMILR is observed in unstable atherosclerotic plaques, and plasma from patients with high plasma C-reactive protein <ref name="ref1" />. | + | ''SMILR'' is expressed highest in heart <ref name="ref1" />. It is differentially expressed in human saphenous vein-derived smooth muscle cells following stimulation with interleukin-1α and platelet-derived growth factor <ref name="ref1" />. Increased expression of ''SMILR'' is observed in unstable atherosclerotic plaques, and plasma from patients with high plasma C-reactive protein <ref name="ref1" />. |
| − | + | ''SMILR'' expression is dysregulated in unstable human carotid plaques <ref name="ref1" />. | |
| − | SMILR | ||
===Regulation=== | ===Regulation=== | ||
| − | SMILR expression can be regulated by IL1α or PDGF and it is a time-dependent manner <ref name="ref1" />. | + | ''SMILR'' expression can be regulated by IL1α or PDGF and it is a time-dependent manner <ref name="ref1" />. |
| + | |||
| + | ===Disease=== | ||
| + | * Cardiovascular disease | ||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
| Line 29: | Line 31: | ||
==References== | ==References== | ||
<references> | <references> | ||
| − | <ref name="ref1"> Ballantyne, M.D., et al. | + | <ref name="ref1"> Ballantyne, M.D., Pinel, K., Dakin, R., Vesey, A.T., Diver, L., Mackenzie, R., Garcia, R., Welsh, P., Sattar, N., Hamilton, G. et al. (2016) Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates Cell Proliferation. Circulation, 133, 2050-2065. |
</ref>(1) | </ref>(1) | ||
</references> | </references> | ||
| + | |||
| + | ===Sequence=== | ||
| + | >gi|105375734|ref|NR_131202.1| Homo sapiens smooth muscle induced lncRNA, enhancer of proliferation (SMILR), long non-coding RNA | ||
| + | <dnaseq>AAATTCTGTTTCGGTTGCAGTTCCATTGTGTGGAGCCAGTACTACAACTTTTCCTCCTGCATTCATTTCT | ||
| + | CTTCCTGCTGCCTTGTGAAGAAGGTGCCTTGCTTCCCCTTCACCTTCTACGGTGATTGCAAGTTGCCTGA | ||
| + | GGATTCCCCAGCCATGTGAAACTTTCACAGAGTTTGAGAGAAACTGTATTCAAGTTGCTGAAACCAAGAA | ||
| + | GCTACACTCACGAGTCTCACCTAAACTCGAATCTGATTTAGATGACATCATCCTGGACTTTGAGTTGATG | ||
| + | AAACCTTGGAGGTCTTGGGAGTAAAGCAAGTGTGATTTGCATATGATGGATATGAATTGTAATGGCCAGA | ||
| + | GCATGGCTGTGGGAACCAAGACTGAATTCCATAATTTACATGGATGTTTGGAAGGTGTCTGCAACTTAAT | ||
| + | CTGTGTTCGTTTCTGAGATGTTGGGCAACTCCTTCTTGGAAGATGTGGTAATGGTCTCTTCGAAAAGAAA | ||
| + | ATAATCATCTGAGTTTTGGCCAAAATAGTTGATCGGATTACCTATGAAAATGACTCTCACCCAACTACAA | ||
| + | GAATGTTAT</dnaseq> | ||
Latest revision as of 01:56, 26 November 2018
SMILR (smooth muscle induced lncRNA, enhancer of proliferationas) is a long noncoding RNA that is used as a driver of vascular smooth muscle cell proliferation and consider as a novel therapeutic strategy to reduce vascular pathologies.
Contents
Annotated Information
Name
SMILR: Smooth muscle induced lncRNA, enhancer of proliferation (HGNC nomenclature), RP11-94a24.1 [1]
Characteristics
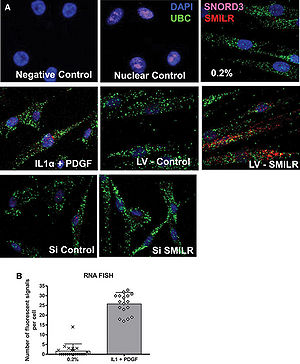
SMILR is four exon transcript of 569 bp long noncoding RNA that is located on human chromosome 8q24.13, ~750 kbp downstream of HAS2 on the same (reverse) strand and ~350 kbp from ZHX2 and ~750 kbp from HAS2-AS1 on the opposite strand of chromosome 8 [1]. The longest open reading frame(ORF) within SMILR is 57 amino acids [1].
SMILR is located in both nucleus and cytoplasm [1].
Function
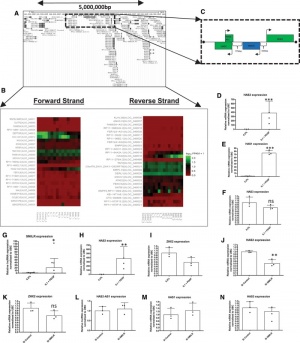
SMILR acts as a driver of vascular smooth muscle cell proliferation [1]. Knockdown of SMILR markedly reduced cell proliferation and HAS2 expression was also reduced by SMILR knockdown [1].
Expression
SMILR is expressed highest in heart [1]. It is differentially expressed in human saphenous vein-derived smooth muscle cells following stimulation with interleukin-1α and platelet-derived growth factor [1]. Increased expression of SMILR is observed in unstable atherosclerotic plaques, and plasma from patients with high plasma C-reactive protein [1].
SMILR expression is dysregulated in unstable human carotid plaques [1].
Regulation
SMILR expression can be regulated by IL1α or PDGF and it is a time-dependent manner [1].
Disease
- Cardiovascular disease
Labs working on this lncRNA
- BHF Glasgow Cardiovascular Research Centre, University of Glasgow, United Kingdom
- British Heart Foundation/University of Edinburgh Centre for Cardiovascular Science, Edinburgh, United Kingdom
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Ballantyne, M.D., Pinel, K., Dakin, R., Vesey, A.T., Diver, L., Mackenzie, R., Garcia, R., Welsh, P., Sattar, N., Hamilton, G. et al. (2016) Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates Cell Proliferation. Circulation, 133, 2050-2065.
Sequence
>gi|105375734|ref|NR_131202.1| Homo sapiens smooth muscle induced lncRNA, enhancer of proliferation (SMILR), long non-coding RNA
000081 CCTTGTGAAG AAGGTGCCTT GCTTCCCCTT CACCTTCTAC GGTGATTGCA AGTTGCCTGA GGATTCCCCA GCCATGTGAA 000160
000161 ACTTTCACAG AGTTTGAGAG AAACTGTATT CAAGTTGCTG AAACCAAGAA GCTACACTCA CGAGTCTCAC CTAAACTCGA 000240
000241 ATCTGATTTA GATGACATCA TCCTGGACTT TGAGTTGATG AAACCTTGGA GGTCTTGGGA GTAAAGCAAG TGTGATTTGC 000320
000321 ATATGATGGA TATGAATTGT AATGGCCAGA GCATGGCTGT GGGAACCAAG ACTGAATTCC ATAATTTACA TGGATGTTTG 000400
000401 GAAGGTGTCT GCAACTTAAT CTGTGTTCGT TTCTGAGATG TTGGGCAACT CCTTCTTGGA AGATGTGGTA ATGGTCTCTT 000480
000481 CGAAAAGAAA ATAATCATCT GAGTTTTGGC CAAAATAGTT GATCGGATTA CCTATGAAAA TGACTCTCAC CCAACTACAA 000560
000561 GAATGTTAT