Difference between revisions of "EPIC1"
| Line 14: | Line 14: | ||
''EPIC1'' promotes breast tumorigenesis through regulating cancer cell-cycle progression <ref name="ref1" />. | ''EPIC1'' promotes breast tumorigenesis through regulating cancer cell-cycle progression <ref name="ref1" />. | ||
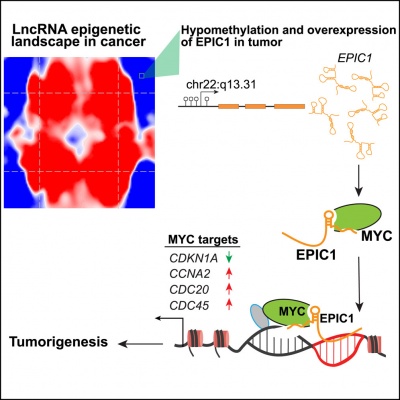
''EPIC1'' interact with MYC and directly regulates the expression of MYC targets through their promoter regions <ref name="ref1" />. ''EPIC1'' may regulate iBET (Bromodomain and Extra-Terminal inhibitors) resistance through increasing MYC protein’s transcriptional activity <ref name="ref2" />. | ''EPIC1'' interact with MYC and directly regulates the expression of MYC targets through their promoter regions <ref name="ref1" />. ''EPIC1'' may regulate iBET (Bromodomain and Extra-Terminal inhibitors) resistance through increasing MYC protein’s transcriptional activity <ref name="ref2" />. | ||
| − | ''EPIC1'' knockdown in MCF-7 cells resulted in the upregulation of 317 genes and downregulation of 488 genes | + | ''EPIC1'' knockdown in MCF-7 cells resulted in the upregulation of 317 genes and downregulation of 488 genes but had little effect on the expression of MYC <ref name="ref1" />. |
''EPIC1'' promotes human lung cancer cell growth by MYC pathway <ref name="ref3" />. | ''EPIC1'' promotes human lung cancer cell growth by MYC pathway <ref name="ref3" />. | ||
''EPIC1'' protects human osteoblasts from dexamethasone (Dex) induced cell death probably through MYC pathway <ref name="ref4" />. | ''EPIC1'' protects human osteoblasts from dexamethasone (Dex) induced cell death probably through MYC pathway <ref name="ref4" />. | ||
[[File:EPIC1.jpg|right|thumb|400px|'''''EPIC1'' interaction with MYC and induced tumerigenesis.''' <ref name="ref1" />]] | [[File:EPIC1.jpg|right|thumb|400px|'''''EPIC1'' interaction with MYC and induced tumerigenesis.''' <ref name="ref1" />]] | ||
| − | |||
| − | |||
===Expression=== | ===Expression=== | ||
''EPIC1'' is overexpressed in luminal breast cancer cell lines (e.g., BT-474, MB361, MCF-7, ZR-75-1, and T-47D), along with ovarian cancer (A2780cis and OVCAR-4), pancreatic cancer (BxPC-3 and PANC-1), prostate cancer (PC-3), and leukemia (K562) cell lines <ref name="ref1" />. | ''EPIC1'' is overexpressed in luminal breast cancer cell lines (e.g., BT-474, MB361, MCF-7, ZR-75-1, and T-47D), along with ovarian cancer (A2780cis and OVCAR-4), pancreatic cancer (BxPC-3 and PANC-1), prostate cancer (PC-3), and leukemia (K562) cell lines <ref name="ref1" />. | ||
''EPIC1'' is upregulated in the necrotic femoral head tissues of Dexamethasone(Dex) taking patients <ref name="ref3" />. | ''EPIC1'' is upregulated in the necrotic femoral head tissues of Dexamethasone(Dex) taking patients <ref name="ref3" />. | ||
| + | |||
| + | ===Regulation=== | ||
| + | ''EPIC1'' is directly controlled by DNA methylation at the CpG islands in its promoter region <ref name="ref1" />. | ||
===Diseases=== | ===Diseases=== | ||
Latest revision as of 06:15, 26 November 2018
EPIC1 interacts with MYC and regulates cell growth and drug-resistance by MYC pathway.
Contents
Annotated Information
Name
EPIC1: Epigenetically induced lncRNA1 [1] LOC284930 (GenBank).
Characteristics
EPIC1 is an intergenic 1815 bp long non-coding RNA, located on chromosome chr22:q13.31, consisting nine exons (GenBank). CpG island is present downstream of EPIC1 gene transcription start site.EPIC1 RNA is predominately located in the nucleus [1].
Function
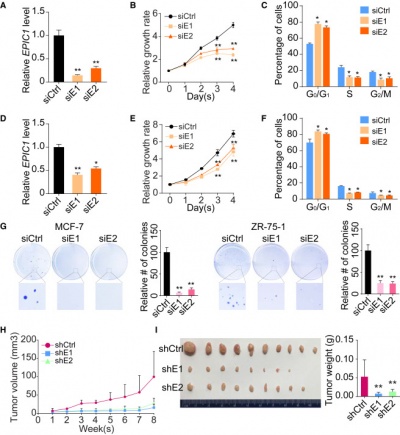
EPIC1 has beed reported to interact with MYC and regulate cell growth [1][2] and drug-resistance [3][4] by MYC pathway. EPIC1 promotes breast tumorigenesis through regulating cancer cell-cycle progression [1]. EPIC1 interact with MYC and directly regulates the expression of MYC targets through their promoter regions [1]. EPIC1 may regulate iBET (Bromodomain and Extra-Terminal inhibitors) resistance through increasing MYC protein’s transcriptional activity [3]. EPIC1 knockdown in MCF-7 cells resulted in the upregulation of 317 genes and downregulation of 488 genes but had little effect on the expression of MYC [1]. EPIC1 promotes human lung cancer cell growth by MYC pathway [2]. EPIC1 protects human osteoblasts from dexamethasone (Dex) induced cell death probably through MYC pathway [4].
Expression
EPIC1 is overexpressed in luminal breast cancer cell lines (e.g., BT-474, MB361, MCF-7, ZR-75-1, and T-47D), along with ovarian cancer (A2780cis and OVCAR-4), pancreatic cancer (BxPC-3 and PANC-1), prostate cancer (PC-3), and leukemia (K562) cell lines [1]. EPIC1 is upregulated in the necrotic femoral head tissues of Dexamethasone(Dex) taking patients [2].
Regulation
EPIC1 is directly controlled by DNA methylation at the CpG islands in its promoter region [1].
Diseases
Labs working on this lncRNA
- Center for Pharmacogenetics, Department of Pharmaceutical Sciences, University of Pittsburgh, Pittsburgh, PA 15261, USA [1]
- University of Pittsburgh Cancer Institute, University of Pittsburgh, Pittsburgh, PA 15261, USA.[1]
- Department of Computational and Systems Biology, University of Pittsburgh, Pittsburgh, PA 15261, USA.[1]
- Department of Respiration, Taizhou People's Hospital, Taizhou, China.[2]
- Department of Oncology, The Fourth Affiliated Hospital of Anhui Medical University, Hefei, China.[2]
- Department of Orthopedics, The Second Affiliated Hospital, Soochow University, Suzhou, China.[4]
- Department of Orthopedics, Tongren Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.[4]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer[J]. Cancer Cell. 2018, 33(4):706-720.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Zhang B, Lu H-Y, Xia Y-H, Jiang A-G & Lv Y-X. Long non-coding RNA EPIC1 promotes human lung cancer cell growth[J]. Biochemical and biophysical research communications. 2018, 503(3):1342-1348.
- ↑ 3.0 3.1 Wang Y, Wang Z, Xu J, Li J, Li S, Zhang M et al. Systematic identification of non-coding pharmacogenomic landscape in cancer[J]. Nature communications. 2018, 9(1):3192.
- ↑ 4.0 4.1 4.2 4.3 Zhang XY, Shan HJ, Zhang P, She C & Zhou XZ. LncRNA EPIC1 protects human osteoblasts from dexamethasone-induced cell death[J]. Biochemical and biophysical research communications. 2018, 503(4):2255-2262.
Sequence
>gi|284930|ref|NR_122046.1| Homo sapiens , EPIC1 long non-coding RNA
000081 CCGGAAGTCA CGTACCCACT CTGTAGGTGC CCCGGGGCAC AGGCAAGCGG ACGAGCCAGT TATCCCTCAG AGCTCCTGCT 000160
000161 GCCTCGCCCG CTTTCTCTCG GAAACGTGAA GTGTGGCCTC AGCTGAAAGT GAGGTGGGCC TCATTCAATC AGTTGAATTC 000240
000241 TTCAAGAGAG AAAAACTGAA GTCCCTTAGA AGGAAAGAGT TCTGCCTTCA GACTGTCTTT GAACTTAAGA CTGTAGCGTC 000320
000321 GACTCCTGCC GGAATTTCCA GCCTGCTGGC CAGCTCTGCA GATTCACACT TGCCAGCCTC CACAATCGCA GCTGAGGCGG 000400
000401 AGGAACCCTA AGGGCTCATT GAGATCATGG ATTTGCCCTT CTATGCATTG ATGGAGCACC TGCTGCCCAC AGCGTCTGTA 000480
000481 TTTGGTGCTG GGATGCTGAG GACACTGAGA TCTTCAAACA GAGGCTGCCA CTCTAAGCAA ACAGATCCCG AGCCCTGGAC 000560
000561 TCTGAAGCTT GGGCCCAGTT CTCCTTTTCT CCGGGTTTCA GATCCCACTG TGAAGTGAGG GGGCCCTTCT GATTCAGGAC 000640
000641 CCGGGGAAGC CAGGGGCATG AGCATCGGTG CCTCTTCTCT ATTTCAAGGA CCCTTCTGGG TGTAAAGTTC TCTGAGATGC 000720
000721 CTTACATGGA TTCCCACCAC TGCAAGATAA CCATCGTATG TAAAGTGTTA TGACCAGCAG AGCGTAATTG AAGTGCATTC 000800
000801 CAGAGGGAAA GACAGCGGCT CAGATGAGGC TTCAGGCTTG ACCAAATCTG ACTGGCTGGC TGCCGGAGAC ACTGCTACCC 000880
000881 ATCGGCCCTT AATAAATGGC CTCATTTGAT CATCATGTGA TCTTCAGAAG CTGGGCTTGA TTATCCTTGC CAAACAAAAT 000960
000961 GCTGAAACCA AAAATCTGAA AGTAAATCAA AGGTTTCGCC ATCATGTAAA CCCTTGGGCG ACAGAGTCTA CCTGTCTCCC 001040
001041 CCTGATATTG TTCTATGAGA TTTCGATGCT ATCTTCATTG CCAGCAGCCC TGACAAAGAT GCACTGCAGA CGGATAGCCT 001120
001121 CTCAGGAACC TGCTTACCTC AGAGCCTGCT GCGGACTGAG TTGTCTTCTC CAAAATCCAT TTGTGGAGGC CCTCATCCCC 001200
001201 AGTGGGAGAG TATTTGGAGA TGAAGCATTT GGAGGTGATT AAGCTTAGAT GAGCTCAGAG TGGAGCCTCA GGATGGCATC 001280
001281 GATGCCCTGG TAAAGAGGGA CACTGAGACC TGGCCCTCCC TTGCTCTCCA CCGTGTGAGG ACACAATGAG ATGGTGGCCA 001360
001361 CCTACAAGCC AGGAACTGAA CCCTACGGGG CCTTGACACT GGCCGTTCCA GCCTCCAGAA TGGCTAGGAA TAAATGTCTG 001440
001441 CTCTTTAAGT CACTGGGCGG TGGAGCTTTG TTATGGCAGC ACCAGGGGCT GGTTGAGAGC CTTTGCATGG CTCTTGTTCC 001520
001521 ACCTGGAATG AGATTTCTAA CTCTTGGCTA CCTCACCCCC TTACCTCATT GTTTCATGTT GCAACCACCC ACCCACTCCC 001600
001601 CATGTCTCTG GCATTCTCAT TTATTTTTTC TTTGATTTCA TAGAACACAT CACAAAGCCA TTTGCTCATG CATGATGTTT 001680
001681 ATTGTCTATT TTCTATCTCC CTGCTAGAGG GTAAGATGCT CAAGAACACT GCTCTGTCTG CCTCCCTGGA CCCAGAAAAT 001760
001761 GCTTGGCGTG TGGTAGAACT TAGATATATT TGTGTTGAAT AAATTAATTG AAGGG