Difference between revisions of "TRPM2-AS"
| Line 18: | Line 18: | ||
[[File: Identification of an antisense transcript and related molecules within the human TRPM2 gene on chromosome 21.jpg|right|thumb|400px|'''Identification of an antisense transcript and related molecules within the human TRPM2 gene on chromosome 21.''' <ref name="ref1" />.]] | [[File: Identification of an antisense transcript and related molecules within the human TRPM2 gene on chromosome 21.jpg|right|thumb|400px|'''Identification of an antisense transcript and related molecules within the human TRPM2 gene on chromosome 21.''' <ref name="ref1" />.]] | ||
[[File: Expression analysis of TRPM2-AS.png|right|thumb|400px|'''Expression analysis of TRPM2-AS.''' <ref name="ref1" />.]] | [[File: Expression analysis of TRPM2-AS.png|right|thumb|400px|'''Expression analysis of TRPM2-AS.''' <ref name="ref1" />.]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
===Characteristics=== | ===Characteristics=== | ||
| Line 34: | Line 28: | ||
TRPM2-AS transcripts is up-regulated in melanoma, and their activation is consistent with the methylation status of the shared CpG island.<ref name="ref1" /> | TRPM2-AS transcripts is up-regulated in melanoma, and their activation is consistent with the methylation status of the shared CpG island.<ref name="ref1" /> | ||
Tumor samples displayed higher levels of TRPM2-AS and TRPM2-TE transcripts with respect to their matched normal tissue. <ref name="ref1" /> | Tumor samples displayed higher levels of TRPM2-AS and TRPM2-TE transcripts with respect to their matched normal tissue. <ref name="ref1" /> | ||
| + | |||
| + | ===Function=== | ||
| + | lncRNA TRPM2-AS possibly regulated cell cycle, apoptosis, and resistance to anti-tumor drugs of carcinoma cells via suppressing p53-p66shc pathway in NSCLC.<ref name="ref2" /> | ||
| + | ===Disease=== | ||
| + | * melanoma<ref name="ref1" /> | ||
| + | * Non-small cell lung cancer (NSCLC) <ref name="ref2" /> | ||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
* DIBIT-San Raffaele Scientific Institute, Via Olgettina 58, 20132 Milan, Italy. <ref name="ref1" /> | * DIBIT-San Raffaele Scientific Institute, Via Olgettina 58, 20132 Milan, Italy. <ref name="ref1" /> | ||
| + | * Respiratory Department, The First Affiliated Hospital of Chinese PLA General Hospital, Beijing, China.<ref name="ref2" /> | ||
==References== | ==References== | ||
| Line 42: | Line 43: | ||
<ref name="ref1">Orfanelli U, Wenke AK, Doglioni C, Russo V, Bosserhoff AK, Lavorgna G.Identification of novel sense and antisense transcription at the TRPM2 locus in cancer. Cell Res. 2008 Nov;18(11):1128-40. doi: 10.1038/cr.2008.296. | <ref name="ref1">Orfanelli U, Wenke AK, Doglioni C, Russo V, Bosserhoff AK, Lavorgna G.Identification of novel sense and antisense transcription at the TRPM2 locus in cancer. Cell Res. 2008 Nov;18(11):1128-40. doi: 10.1038/cr.2008.296. | ||
</ref>(1) | </ref>(1) | ||
| + | |||
| + | <ref name="ref2">Ma LY, Xie XW, Ma L, et al. Downregulated long non-coding RNA TRPM2-AS inhibits cisplatin resistance of non-small cell lung cancer cells via activation of p53- p66shc pathway[J]. Eur Rev Med Pharmacol Sci, 2017, 21: 2626-2634. | ||
| + | </ref>(2) | ||
</references> | </references> | ||
Latest revision as of 14:49, 10 August 2019
Contents
Annotated Information
Approved Symbol
Name
Approved symbol: TRPM2-AS
Approved name: TRPM2 antisense RNA
HGNC ID: HGNC:50758
Alias symbols: TRPM2-AS1
RefSeq ID: NR_109964
Ensembl ID: ENSG00000230061
LncBook ID: HSALNT0289414

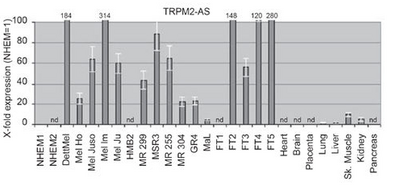
Characteristics
TRPM2-AS is antisense with respect to TRPM2, located at chromosome 21q22.3[2], an ion channel capable of conferring cell death upon oxidative stress.[1]
Regulation
The CpG island could be involved in the transcriptional regulation of TRPM2-AS,specifically,the methylation status of the CpG island contributes negatively to the expression level of both TRPM2-AS.[1]
Expression
TRPM2-AS transcripts is up-regulated in melanoma, and their activation is consistent with the methylation status of the shared CpG island.[1] Tumor samples displayed higher levels of TRPM2-AS and TRPM2-TE transcripts with respect to their matched normal tissue. [1]
Function
lncRNA TRPM2-AS possibly regulated cell cycle, apoptosis, and resistance to anti-tumor drugs of carcinoma cells via suppressing p53-p66shc pathway in NSCLC.[2]
Disease
Labs working on this lncRNA
- DIBIT-San Raffaele Scientific Institute, Via Olgettina 58, 20132 Milan, Italy. [1]
- Respiratory Department, The First Affiliated Hospital of Chinese PLA General Hospital, Beijing, China.[2]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Orfanelli U, Wenke AK, Doglioni C, Russo V, Bosserhoff AK, Lavorgna G.Identification of novel sense and antisense transcription at the TRPM2 locus in cancer. Cell Res. 2008 Nov;18(11):1128-40. doi: 10.1038/cr.2008.296.
- ↑ 2.0 2.1 2.2 2.3 Ma LY, Xie XW, Ma L, et al. Downregulated long non-coding RNA TRPM2-AS inhibits cisplatin resistance of non-small cell lung cancer cells via activation of p53- p66shc pathway[J]. Eur Rev Med Pharmacol Sci, 2017, 21: 2626-2634.
Sequence
>gi|568786345|ref|NR_109964.1| Homo sapiens TRPM2 antisense RNA (TRPM2-AS), long non-coding RNA
000081 CGGGAGCTCA GGCAGCGTCC CCGCCGGCAC CTGCCTCTTT GTGCAGCGGC AGTCAGGGCC CTCGGCAATG AGCTGAACTC 000160
000161 GCAGTGCGGT TACGAGGGCA AATATGCTCC TTGAGGGCCG GCGAAACTCA GCAGCCCGAG GAAGGCTACT GATGTGCATT 000240
000241 TCATAGCCGG CTGCGAATTT AGGAAAACAG ACTTTGCTTC TCGTCACTCA GCCTACGTGA CCAGGTTCAG ACACAGTCTG 000320
000321 CAGCCGCCCG CCTCGCACCC CCACTCTGCA GTGCGGTATG TGGGGAGCTC AGGGCACAGC AGGCCAGGCC TCCCCGTGGA 000400
000401 CGTCCAGGAA CCAGAACCAA ACTGCCCAGG GCCCCAGAGG GGAAGATGTC TCAGCAGACG GACAGCCGAG GCTCACATGG 000480
000481 CAAGCTCTGG CAGCCTGTCG GTCCCAGGAG AGAGGGGGAG ATGGCAGACG GGAAAAGAAG CCACCTGCTG GGATGCTGAG 000560
000561 ACTCGCTTGC AGGAGCTTTT GGAACTGGCT GAGGTCACAG CTGGAACCAC TGTGGCCAGC TGGAGTCTGC ACAGCCCGAG 000640
000641 TTTCCACCCC AGGGTCCCAG TAACTCCGCC CAAATGTGCA CACGAGACCT ATGAGGAGAC ATAACTTTCC AGAACCCCCT 000720
000721 TTTCTTCCAC CAGCCACTTA CTCATCCAAG AACCCACCCC CGAACCTTCC CTAATAGAAA CACTGCATTA AAGCCAGCGC 000800
000801 GGGGAGACAG ACGTGAACTG CGCCCCTGTC TCCTTGTGGG TTGGCCTAGA ATAAAAGCTT TTCTTTTCTC AAAAA