Difference between revisions of "NONHSAT001953"
Qianpeng Li (talk | contribs) |
|||
| (5 intermediate revisions by 4 users not shown) | |||
| Line 4: | Line 4: | ||
===Name=== | ===Name=== | ||
SNHG3: Small nucleolar RNA host gene 3 | SNHG3: Small nucleolar RNA host gene 3 | ||
| + | |||
| + | Previous symbols: RNU17C | ||
| + | |||
| + | Alias symbols: U17HG; U17HG-A; NCRNA00014 | ||
| + | |||
| + | HGNC ID : HGNC:10118 | ||
| + | |||
| + | RefSeq ID: NR_002909.1 | ||
| + | |||
| + | ===Disease=== | ||
| + | Alzheimer's disease | ||
| + | |||
| + | Glioma[http://www.ncbi.nlm.nih.gov/pubmed/30042166 (Fei (2018))] | ||
===Characteristics=== | ===Characteristics=== | ||
| − | Upstream from the RCC1 protein coding gene. Multiple SNHG3 splice isoforms exist, minority join RCC1, majority do not. Isoforms range from ~0.9kb to ~2.6kb in human. | + | Upstream from the RCC1 protein-coding gene. Multiple SNHG3 splice isoforms exist, minority join RCC1, majority do not. Isoforms range from ~0.9kb to ~2.6kb in human. |
Contain U17a and U17b snoRNAs in introns. | Contain U17a and U17b snoRNAs in introns. | ||
| − | SNHG3 contains a number of exonic Alu elements | + | SNHG3 contains a number of exonic Alu elements. |
Transcriptional start site has characteristics of 5' TOP gene family. | Transcriptional start site has characteristics of 5' TOP gene family. | ||
| + | |||
| + | Cell fractionation experiments indicated that U17HG RNA is enriched in the cytoplasm[http://www.ncbi.nlm.nih.gov/pubmed/9671460 (Pelczar (1998))]. | ||
===Function=== | ===Function=== | ||
| − | Involved in maintaining pluripotency in ESCs [http://www.ncbi.nlm.nih.gov/pubmed/21874018 (Guttman (2011))]. | + | * Involved in maintaining pluripotency in ESCs [http://www.ncbi.nlm.nih.gov/pubmed/21874018 (Guttman (2011))]. |
| − | Knockdown in mECSs lead to down-regulation of Nanog, Sox2, Klf4 and Oct4 expression as well as changes to ESC morphology [http://www.ncbi.nlm.nih.gov/pubmed/21874018 (Guttman (2011))]. | + | * Knockdown in mECSs lead to down-regulation of Nanog, Sox2, Klf4 and Oct4 expression as well as changes to ESC morphology [http://www.ncbi.nlm.nih.gov/pubmed/21874018 (Guttman (2011))]. |
| − | Snhg3 was found to interact with a number of chromatin binding proteins/complexes in mESCs including PRC1, PRC2, JARID1B and SUV39H1, with the general pattern being interaction with repressors of gene expression [http://www.ncbi.nlm.nih.gov/pubmed/21874018 (Guttman (2011))]. | + | * Snhg3 was found to interact with a number of chromatin binding proteins/complexes in mESCs including PRC1, PRC2, JARID1B and SUV39H1, with the general pattern being interaction with repressors of gene expression [http://www.ncbi.nlm.nih.gov/pubmed/21874018 (Guttman (2011))]. |
| + | * The up-regulation of SNHG3 promoted cell proliferation, accelerate cell cycle progress, and repressed cell apoptosis. Mechanically, SNHG3 facilitated the malignant progression of glioma through epigenetically repressing KLF2 and p21 via recruiting enhancer of zeste homolog 2 to the promoter of KLF2 and p21[http://www.ncbi.nlm.nih.gov/pubmed/30042166 (Fei (2018))]. | ||
===Expression=== | ===Expression=== | ||
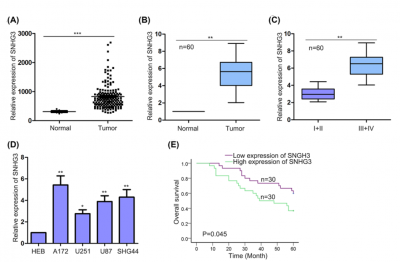
| + | [[File:SNHG3exp.png|right|thumb|400px|The expression pattern of SNHG3 in glioma tissues and normal tissues[http://www.ncbi.nlm.nih.gov/pubmed/30042166 (Fei (2018))].]] | ||
| + | |||
Localised to cytoplasm. Unlike SNHG1 it was not stabilised by translation inhibitors or associated with translating ribosomes and was unaffected by translation inhibitors. | Localised to cytoplasm. Unlike SNHG1 it was not stabilised by translation inhibitors or associated with translating ribosomes and was unaffected by translation inhibitors. | ||
| Line 28: | Line 46: | ||
Transcript found to be unstable with a half-life <2 hr in mouse N2A (neuroblastoma) cells and human B cells ([http://www.ncbi.nlm.nih.gov/pubmed/19561200 Friedel (2009)], [http://www.ncbi.nlm.nih.gov/pubmed/22406755 Clark (2012)]). | Transcript found to be unstable with a half-life <2 hr in mouse N2A (neuroblastoma) cells and human B cells ([http://www.ncbi.nlm.nih.gov/pubmed/19561200 Friedel (2009)], [http://www.ncbi.nlm.nih.gov/pubmed/22406755 Clark (2012)]). | ||
| + | SNHG3 was more highly expressed in glioma tissues, especially in tissues with advanced tumor grade, and cell lines[http://www.ncbi.nlm.nih.gov/pubmed/30042166 (Fei (2018))]. | ||
| + | {| class='wikitable' style="text-align:center" | ||
| + | |- | ||
| + | ! | Experiment | ||
| + | ! | Forward primer | ||
| + | ! | Reverse primer | ||
| + | |- | ||
| + | | rowspan="1"|Quantitative real-time PCR | ||
| + | | | 5'-TTCAAGCGATTCTCGTGCC- 3' | ||
| + | | | 5'-AAGATTGTCAAACCCTCCCTGT- 3'[http://www.ncbi.nlm.nih.gov/pubmed/30042166 (Fei (2018))] | ||
| + | |} | ||
===Conservation=== | ===Conservation=== | ||
Not highly conserved, ~40-50% identity between mouse and human exons. | Not highly conserved, ~40-50% identity between mouse and human exons. | ||
| Line 68: | Line 97: | ||
sequence = <dnaseq>GATTCTCTAACTGCGCATGCTTCTGCGCACGCGCAATAGACATTCCAGGACTTCCGGGCACTTCGTAAGGTTTAAAAAGGATGCTTCGCGTTTTCTCTCTCCTTTTTGGAGACAGATTCGCAGTGGTCGCTTCTTCTCCTTGACGGAGTCGGTTTGTCACTCAGGCTGGACTGCAGTGCTCGTTGCAACCTCCGCCTGCCGGGTTCAAGCGATTCTCGTGCCTCAGCCTCTCCAGCAGCTGGGATTACAGGATTTGTTAAGGATTCCAAGTAACTCTTATTTGGAGAGAAGACGATCTGCACTTCGCATTTTGGCATTGACATTTAATTTTAGGGTCCTTTATATAGAAGGGAGAGTAGGTAAACTGATTTTTTTTTTTAACAGGGAGGGTTTGACAATCTTTGGCAGACTTGGAGCAAAAGATTGAGGTGCATTTCATGCCTCCTTTTGAGAGTCTTGCTCTGTCGCCCAGGCTGTAGTGCAGTGGCGCAATCTTGGCTGCAACCTCAGCCTCCCAAGTAGCTGGGATTACAAACATAAGCCACCACGCCCAGCCCTCATACCTCTTTTAAAAGTCGACCTGTTTTGCAGAAAGTCTGCTGTTTTTGTACTAAAGGCTTTGGAATTTGGCATTTAGCTAGGAATGCACATTCTTTCACCTCATTCATACTTTAAGAACCACAGAAGTGACTCTGCTTGGCCAGAAGGCACACTGTGTTGGTGGTTATATTAAAAGTCCTTGAGTATTTTGCTTTTCATGATCTTGCTCACTGCAACTTCCGCCTCCCAGGTTCAGGCGATTCTCCTGCCTCAGCCTCCCAAGTAGCTGCGACTACAGGCGTGTAGCACCACACCTGGCTAATTTTTGTATTTTTAGTAGAGATGAGGTTTCACCATATTGGCCAGGCTGTTCTCAACTCCTGACCTCGTGATCCGCCCACCTCAGCCTCCTAAAGTGCTGGGATTACAGCTGTGAGCCACCCTGCCCGGCCACTTTTGTATGATTTCTAATGTATTTGTAATTTACCTAACAAATTGCCTAATCTGCTATGTTAATGTATTTATGAATTAAAATAAATACGACTGCATGTTTGTGGTTCATTTTTGTGGAGGTGGCTGTGGTGACATCAGCCAAGAATCTGAATGGTACTGTTGAAGGAAACTAGCATGATAGCTTCAGTTCTAAAGGCCCTGAAACCTAGTCTCAGGTGGGTCCCCCTTGGGTTCACTTTATATTGGCAGTTTATTGGGAAAATGGATATTAGGTCCTGACCAATAGGACCGTAAGTCTGGGTTGAGTGCAAGATGAGTTAGACCGATTCTTTAGCTTCCTGCAGTGTAGTGGAGGAAAAATCGATGGTAGCAACGGGAGGTTGTATCCCTAGCTGATGAGTTGTATGAGCCTCTACTACCTGGCGCACCTCCGCCTGAAGATTGCCAGAATTGCTTGCCTCATGACGTGAGTCACAATGGAAACTTTGTCAAGCCCCCTGCACTGGCTGCCAACATAAATGTTCAGTACCCTGAAGGATGGGACTGAAGGGGGATCATCTAGAAGGTAAAGTTACCTACTGGCATAGGGGAGGTGGGACAGCCGTTAAGCCATTTGGAACTTGATGGAGACAGGTTTGAGGGAGGTGGGTGAGATTGGAGTTTGGTGGACTGTAGAGCTTGCTTGCCAAGGTGTTGAGGTCAGGGTTGGTTTGAGAATGGAAGCTAGTTACTAGCTATGATTGTGGGGGAACACAGCTTGATTTTTCTTACAAGCTAAGAGGAGTGAGGCAGTGTTTAAGAGGGCATGTTAAATGCAGCCAGGCTTGGTGGCTCACACCCGTAATCCCAGCACTTAGGCTAAGGCAGGCGGATCACAACATCTAGAGATCCTGGCCAACGCGGTGAAACCCTGTCTGTACTAAAAATACAAAATAACTGGGCATGGTGGTGTGCACCTGTGGGAGGCTGAGGCAGAATTGCTGGAACCCGGGAGATGGAGGTTGTACTGAGCTGAGACCTTGCCACTGCGCTCCAGCCTGGTGACAGAGTTAAGTCTCAAAAAAAAGGCATCTTCCTAAAGCAATTGTATTTGTGCTTACCTGTGCCAGGCACTGTTCTAGGTAAGCACTAAGTGGGCTTTAATACAGCATATTCCAATGGGGAATCCCAGGAACCAAAAGACTAATTGTCCAAGTCCACAACTAGAAGTGGCACCTCTGCAGAAACAAGCATCAAATTCCCTGCTCAGGAAGAAGCCAGATGAGTCAGCCCCATTCGTCTGTATGCCCAGTCCCATCCGTGTCCTGCTGTAACTACATAGATCTCACCTGAGTAAAGTGATTTTTTTCTGAA</dnaseq>| | sequence = <dnaseq>GATTCTCTAACTGCGCATGCTTCTGCGCACGCGCAATAGACATTCCAGGACTTCCGGGCACTTCGTAAGGTTTAAAAAGGATGCTTCGCGTTTTCTCTCTCCTTTTTGGAGACAGATTCGCAGTGGTCGCTTCTTCTCCTTGACGGAGTCGGTTTGTCACTCAGGCTGGACTGCAGTGCTCGTTGCAACCTCCGCCTGCCGGGTTCAAGCGATTCTCGTGCCTCAGCCTCTCCAGCAGCTGGGATTACAGGATTTGTTAAGGATTCCAAGTAACTCTTATTTGGAGAGAAGACGATCTGCACTTCGCATTTTGGCATTGACATTTAATTTTAGGGTCCTTTATATAGAAGGGAGAGTAGGTAAACTGATTTTTTTTTTTAACAGGGAGGGTTTGACAATCTTTGGCAGACTTGGAGCAAAAGATTGAGGTGCATTTCATGCCTCCTTTTGAGAGTCTTGCTCTGTCGCCCAGGCTGTAGTGCAGTGGCGCAATCTTGGCTGCAACCTCAGCCTCCCAAGTAGCTGGGATTACAAACATAAGCCACCACGCCCAGCCCTCATACCTCTTTTAAAAGTCGACCTGTTTTGCAGAAAGTCTGCTGTTTTTGTACTAAAGGCTTTGGAATTTGGCATTTAGCTAGGAATGCACATTCTTTCACCTCATTCATACTTTAAGAACCACAGAAGTGACTCTGCTTGGCCAGAAGGCACACTGTGTTGGTGGTTATATTAAAAGTCCTTGAGTATTTTGCTTTTCATGATCTTGCTCACTGCAACTTCCGCCTCCCAGGTTCAGGCGATTCTCCTGCCTCAGCCTCCCAAGTAGCTGCGACTACAGGCGTGTAGCACCACACCTGGCTAATTTTTGTATTTTTAGTAGAGATGAGGTTTCACCATATTGGCCAGGCTGTTCTCAACTCCTGACCTCGTGATCCGCCCACCTCAGCCTCCTAAAGTGCTGGGATTACAGCTGTGAGCCACCCTGCCCGGCCACTTTTGTATGATTTCTAATGTATTTGTAATTTACCTAACAAATTGCCTAATCTGCTATGTTAATGTATTTATGAATTAAAATAAATACGACTGCATGTTTGTGGTTCATTTTTGTGGAGGTGGCTGTGGTGACATCAGCCAAGAATCTGAATGGTACTGTTGAAGGAAACTAGCATGATAGCTTCAGTTCTAAAGGCCCTGAAACCTAGTCTCAGGTGGGTCCCCCTTGGGTTCACTTTATATTGGCAGTTTATTGGGAAAATGGATATTAGGTCCTGACCAATAGGACCGTAAGTCTGGGTTGAGTGCAAGATGAGTTAGACCGATTCTTTAGCTTCCTGCAGTGTAGTGGAGGAAAAATCGATGGTAGCAACGGGAGGTTGTATCCCTAGCTGATGAGTTGTATGAGCCTCTACTACCTGGCGCACCTCCGCCTGAAGATTGCCAGAATTGCTTGCCTCATGACGTGAGTCACAATGGAAACTTTGTCAAGCCCCCTGCACTGGCTGCCAACATAAATGTTCAGTACCCTGAAGGATGGGACTGAAGGGGGATCATCTAGAAGGTAAAGTTACCTACTGGCATAGGGGAGGTGGGACAGCCGTTAAGCCATTTGGAACTTGATGGAGACAGGTTTGAGGGAGGTGGGTGAGATTGGAGTTTGGTGGACTGTAGAGCTTGCTTGCCAAGGTGTTGAGGTCAGGGTTGGTTTGAGAATGGAAGCTAGTTACTAGCTATGATTGTGGGGGAACACAGCTTGATTTTTCTTACAAGCTAAGAGGAGTGAGGCAGTGTTTAAGAGGGCATGTTAAATGCAGCCAGGCTTGGTGGCTCACACCCGTAATCCCAGCACTTAGGCTAAGGCAGGCGGATCACAACATCTAGAGATCCTGGCCAACGCGGTGAAACCCTGTCTGTACTAAAAATACAAAATAACTGGGCATGGTGGTGTGCACCTGTGGGAGGCTGAGGCAGAATTGCTGGAACCCGGGAGATGGAGGTTGTACTGAGCTGAGACCTTGCCACTGCGCTCCAGCCTGGTGACAGAGTTAAGTCTCAAAAAAAAGGCATCTTCCTAAAGCAATTGTATTTGTGCTTACCTGTGCCAGGCACTGTTCTAGGTAAGCACTAAGTGGGCTTTAATACAGCATATTCCAATGGGGAATCCCAGGAACCAAAAGACTAATTGTCCAAGTCCACAACTAGAAGTGGCACCTCTGCAGAAACAAGCATCAAATTCCCTGCTCAGGAAGAAGCCAGATGAGTCAGCCCCATTCGTCTGTATGCCCAGTCCCATCCGTGTCCTGCTGTAACTACATAGATCTCACCTGAGTAAAGTGATTTTTTTCTGAA</dnaseq>| | ||
}} | }} | ||
| − | [[Category:Intergenic]] | + | [[Category:Intergenic]][[Category:NONHSAG000835]][[Category:Transcripts]] |
Latest revision as of 07:06, 1 August 2019
Please input one-sentence summary here.
Contents
Annotated Information
Name
SNHG3: Small nucleolar RNA host gene 3
Previous symbols: RNU17C
Alias symbols: U17HG; U17HG-A; NCRNA00014
HGNC ID : HGNC:10118
RefSeq ID: NR_002909.1
Disease
Alzheimer's disease
Glioma(Fei (2018))
Characteristics
Upstream from the RCC1 protein-coding gene. Multiple SNHG3 splice isoforms exist, minority join RCC1, majority do not. Isoforms range from ~0.9kb to ~2.6kb in human.
Contain U17a and U17b snoRNAs in introns.
SNHG3 contains a number of exonic Alu elements.
Transcriptional start site has characteristics of 5' TOP gene family.
Cell fractionation experiments indicated that U17HG RNA is enriched in the cytoplasm(Pelczar (1998)).
Function
- Involved in maintaining pluripotency in ESCs (Guttman (2011)).
- Knockdown in mECSs lead to down-regulation of Nanog, Sox2, Klf4 and Oct4 expression as well as changes to ESC morphology (Guttman (2011)).
- Snhg3 was found to interact with a number of chromatin binding proteins/complexes in mESCs including PRC1, PRC2, JARID1B and SUV39H1, with the general pattern being interaction with repressors of gene expression (Guttman (2011)).
- The up-regulation of SNHG3 promoted cell proliferation, accelerate cell cycle progress, and repressed cell apoptosis. Mechanically, SNHG3 facilitated the malignant progression of glioma through epigenetically repressing KLF2 and p21 via recruiting enhancer of zeste homolog 2 to the promoter of KLF2 and p21(Fei (2018)).
Expression
Localised to cytoplasm. Unlike SNHG1 it was not stabilised by translation inhibitors or associated with translating ribosomes and was unaffected by translation inhibitors.
Identified as expressed in mouse embryonic stem cells (Guttman (2011)).
Transcript found to be unstable with a half-life <2 hr in mouse N2A (neuroblastoma) cells and human B cells (Friedel (2009), Clark (2012)).
SNHG3 was more highly expressed in glioma tissues, especially in tissues with advanced tumor grade, and cell lines(Fei (2018)).
| Experiment | Forward primer | Reverse primer |
|---|---|---|
| Quantitative real-time PCR | 5'-TTCAAGCGATTCTCGTGCC- 3' | 5'-AAGATTGTCAAACCCTCCCTGT- 3'(Fei (2018)) |
Conservation
Not highly conserved, ~40-50% identity between mouse and human exons.
Misc
Please input misc information here.
Transcriptomic Nomeclature
Please input transcriptomic nomeclature information here.
Regulation
Please input regulation information here.
Allelic Information and Variation
Please input allelic information and variation information here.
Evolution
Please input evolution information here.
You can also add sub-section(s) at will.
Labs working on this lncRNA
Please input related labs here.
References
Please input cited references here.
Basic Information
| Transcript ID |
NONHSAT001953 |
| Source |
NONCODE4.0 |
| Same with |
, |
| Classification |
intergenic |
| Length |
2346 nt |
| Genomic location |
chr1+:28832455..28837404 |
| Exon number |
4 |
| Exons |
28832455..28832596,28832732..28832839,28834640..28834672,28835342..28837404 |
| Genome context |
|
| Sequence |
000001 GATTCTCTAA CTGCGCATGC TTCTGCGCAC GCGCAATAGA CATTCCAGGA CTTCCGGGCA CTTCGTAAGG TTTAAAAAGG 000080 000081 ATGCTTCGCG TTTTCTCTCT CCTTTTTGGA GACAGATTCG CAGTGGTCGC TTCTTCTCCT TGACGGAGTC GGTTTGTCAC 000160 000161 TCAGGCTGGA CTGCAGTGCT CGTTGCAACC TCCGCCTGCC GGGTTCAAGC GATTCTCGTG CCTCAGCCTC TCCAGCAGCT 000240 000241 GGGATTACAG GATTTGTTAA GGATTCCAAG TAACTCTTAT TTGGAGAGAA GACGATCTGC ACTTCGCATT TTGGCATTGA 000320 000321 CATTTAATTT TAGGGTCCTT TATATAGAAG GGAGAGTAGG TAAACTGATT TTTTTTTTTA ACAGGGAGGG TTTGACAATC 000400 000401 TTTGGCAGAC TTGGAGCAAA AGATTGAGGT GCATTTCATG CCTCCTTTTG AGAGTCTTGC TCTGTCGCCC AGGCTGTAGT 000480 000481 GCAGTGGCGC AATCTTGGCT GCAACCTCAG CCTCCCAAGT AGCTGGGATT ACAAACATAA GCCACCACGC CCAGCCCTCA 000560 000561 TACCTCTTTT AAAAGTCGAC CTGTTTTGCA GAAAGTCTGC TGTTTTTGTA CTAAAGGCTT TGGAATTTGG CATTTAGCTA 000640 000641 GGAATGCACA TTCTTTCACC TCATTCATAC TTTAAGAACC ACAGAAGTGA CTCTGCTTGG CCAGAAGGCA CACTGTGTTG 000720 000721 GTGGTTATAT TAAAAGTCCT TGAGTATTTT GCTTTTCATG ATCTTGCTCA CTGCAACTTC CGCCTCCCAG GTTCAGGCGA 000800 000801 TTCTCCTGCC TCAGCCTCCC AAGTAGCTGC GACTACAGGC GTGTAGCACC ACACCTGGCT AATTTTTGTA TTTTTAGTAG 000880 000881 AGATGAGGTT TCACCATATT GGCCAGGCTG TTCTCAACTC CTGACCTCGT GATCCGCCCA CCTCAGCCTC CTAAAGTGCT 000960 000961 GGGATTACAG CTGTGAGCCA CCCTGCCCGG CCACTTTTGT ATGATTTCTA ATGTATTTGT AATTTACCTA ACAAATTGCC 001040 001041 TAATCTGCTA TGTTAATGTA TTTATGAATT AAAATAAATA CGACTGCATG TTTGTGGTTC ATTTTTGTGG AGGTGGCTGT 001120 001121 GGTGACATCA GCCAAGAATC TGAATGGTAC TGTTGAAGGA AACTAGCATG ATAGCTTCAG TTCTAAAGGC CCTGAAACCT 001200 001201 AGTCTCAGGT GGGTCCCCCT TGGGTTCACT TTATATTGGC AGTTTATTGG GAAAATGGAT ATTAGGTCCT GACCAATAGG 001280 001281 ACCGTAAGTC TGGGTTGAGT GCAAGATGAG TTAGACCGAT TCTTTAGCTT CCTGCAGTGT AGTGGAGGAA AAATCGATGG 001360 001361 TAGCAACGGG AGGTTGTATC CCTAGCTGAT GAGTTGTATG AGCCTCTACT ACCTGGCGCA CCTCCGCCTG AAGATTGCCA 001440 001441 GAAT |