Difference between revisions of "NONHSAT087920"
Chunlei Yu (talk | contribs) (→Annotated Information) |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | GHRLOS, which gives rise to endogenous ghrelin natural antisense transcripts. exhibits features which are common to many non-coding RNA genes, including extensive splicing, lack of significant and conserved open reading frames, differential expression and lack of conservation in vertebrates. | |
==Annotated Information== | ==Annotated Information== | ||
| − | === | + | ===Name=== |
| − | + | GHRLOS:ghrelin opposite strand/antisense RNA(HGNC nomenclature) | |
| + | |||
| + | "GHRL antisense RNA 1 (non-protein coding)", GHRL-AS1, NCRNA00068, "non-protein coding RNA 68"<ref name="ref1" /> | ||
| + | |||
| + | ===Characteristics=== | ||
| + | [[File:Mapping of GHRLOS transcript 5' and 3' ends.jpg|right|thumb|400px|'''Mapping of GHRLOS transcript 5' and 3' ends.'''<ref name="ref1" />]] | ||
| + | |||
| + | GHRLOS transcripts that extend over 1.4 kb of the promoter region and 106 nucleotides of exon 4 of the ghrelin gene(GHRL) initiate 4.8 kb downstream of the terminal exon 4 of GHRL and are present in the 3' untranslated exon of the adjacent gene TATDN2 (TatD DNase domain containing 2). The 3' region of GHRLOS is also antisense, in a tail-to-tail fashion to a novel terminal exon of the neighbouring SEC13 gene, which is important inprotein transport. GHRLOS that is riddled with stop codons is little nucleotide and amino-acid sequence conservation between vertebrates. The gene spans 44 kb on 3p25.3, is extensively spliced and harbours multiple variable exons<ref name="ref1" />. | ||
===Function=== | ===Function=== | ||
| − | + | GHRLOS RNA transcripts display several distinctive features of non-coding (ncRNA) genes, including 5' capping, polyadenylation, extensive splicing and short open reading frames<ref name="ref1" />. | |
| + | GHRLOS natural antisense transcripts, functioning as a non-coding RNA,may regulate one or both of these genes, because GHRLOS overlaps the terminal exons of both GHRL andSEC13-T<ref name="ref1" /> | ||
| + | |||
| + | ===Expression=== | ||
| + | We have also investigated the expression of GHRLOS and found evidence of differential tissue expression. It is highly expressed in tissues which are emerging as major sites of non-coding RNA expression (the thymus, brain, and testis), as well as in the ovary and uterus. In contrast, very low levels were found in the stomach where sense, GHRL derived RNAs are highly expressed. | ||
| + | For non-quantitative RT-PCR analysis of GHRLOS splicing, RT-PCRs were performed with a forward primer in a region common to the 5' terminal exon Ia/b and a reverse primer in the 3' terminal exon 4 of GHRLOS<ref name="ref1" />. | ||
| + | |||
| + | {|class='wikitable' style="text-align:center" | ||
| + | |- | ||
| + | | rowspan="1"| Ito4-F | ||
| + | | | CATGGAAGTCTCCAAGCCTG<ref name="ref1" /> | ||
| + | |- | ||
| + | | rowspan="1"| Ito4-R | ||
| + | | | CTGCTCTACTGCCTCAATGTC<ref name="ref1" /> | ||
| + | |} | ||
| + | |||
| + | To detect long, chimaeric transcripts, we employed RT-PCR with a forward primer in exon 2 of TATDN2(ChiOut-F) and a reverse primer in exon 1 of GHRLOS<ref name="ref1" />. | ||
| − | === | + | {|class='wikitable' style="text-align:center" |
| − | + | |- | |
| + | | rowspan="1"| ChiOut-F | ||
| + | | | TGAAAGCCCAGAAGGAGGA<ref name="ref1" /> | ||
| + | |- | ||
| + | | rowspan="1"| ChiOut-R | ||
| + | | | TCTAAGTTTAGAAGCGCTCATCTG<ref name="ref1" /> | ||
| + | |} | ||
| − | = | + | To allow strand-specific and RNA-specific amplification of GHRLOS transcripts, reverse transcription was performed using a gene-specific primer in exon 4 with a linker (LK) sequence attached to the 5' end of the primer<ref name="ref1" />. |
| − | |||
| − | === | + | {|class='wikitable' style="text-align:center" |
| − | + | |- | |
| + | | rowspan="1"| GHRLOS-Real-F | ||
| + | | | CATTGAGGCAGTAGAGCAGTTGA<ref name="ref1" /> | ||
| + | |- | ||
| + | | rowspan="1"| LK | ||
| + | | | CGACTGGAGCACGAGGACACTGA<ref name="ref1" /> | ||
| + | |} | ||
| − | = | + | To detect sense GHRL transcripts,a strand-specific RT-PCR approach was employed, with a reverse transcription primer spanning the 3' terminal exon 4 of the ghrelin gene (GHRLex4_RT_LK) followed by PCR with an exon 4 specific forward primer (GHRLex4_F) and a linker-specific reverse primer(LK)<ref name="ref1" />. |
| − | |||
| − | + | {|class='wikitable' style="text-align:center" | |
| + | |- | ||
| + | | rowspan="1"| GHRL-Real-RT-LK | ||
| + | | | CGACTGGAGCACGAGGACACTGAGCCAGAGAGCGCTTCTAAACTTA<ref name="ref1" /> | ||
| + | |- | ||
| + | | rowspan="1"| GHRL-Real-F | ||
| + | | | GCCCCAGCCGACAAGTG<ref name="ref1" /> | ||
| + | |- | ||
| + | | rowspan="1"| LK | ||
| + | | | CGACTGGAGCACGAGGACACTGA<ref name="ref1" /> | ||
| + | |} | ||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
| − | + | Institute of Health and Biomedical Innovation, Queensland University of Technology, Kelvin Grove, Queensland, Australia<ref name="ref1" />. | |
==References== | ==References== | ||
| − | + | <references> | |
| + | <ref name="ref1">Seim I, Carter SL, Herington AC, Chopin LK. Complex organisation and structure of the ghrelin antisense strand gene GHRLOS, a candidate non-coding RNA gene[J]. BMC molecular biology. 2008,9:95.</ref>(1) | ||
| + | </references> | ||
{{basic| | {{basic| | ||
tID = NONHSAT087920| | tID = NONHSAT087920| | ||
source = NONCODE4.0| | source = NONCODE4.0| | ||
| − | same = ,| | + | same = GHRLOS, GHRL-AS1, NCRNA00068| |
| − | classification = | + | classification = antisense| |
length = 1765 nt| | length = 1765 nt| | ||
location = chr3+:10322636..10335133| | location = chr3+:10322636..10335133| | ||
| Line 42: | Line 87: | ||
sequence = <dnaseq>ACATGGAAGTCTCCAAGCCTGTGCCATCCACCTGCCAAGGAAAAGCACAAGCATACAGTTTGAACATTTATTCGCCTCCTGAGCTTGTACAACAGTCGTGGGAGTTGCTGCAGAAGCAAGCGAAAAGCCAGATGAGCGCTTCTAAACTTAGAGAGAGGGAGAGCGCCTCATCTCTTCCATTTTCCAGGTATTCCTGAGATGATTTATTGGAGCTCAAAGCTTTGGGAGAGTTGGGGCCTTCCATTCCCTCCAGTAAATACTTGTTTTTCTTCCACCGCTGAGGCAAATGCGGGGTGGCTGATCACCTGGCAGACATCTTAGGAAACAGGAGCACCGGTCTGGGAAACTGCTGGCCTGGCCTAACACCTGGCGCTGTGGTGCAGGAGGGCAGCCACTGAGGATTCTTGAGGAGAGAGAGaccatagaatgaatgaaagagttggaaagactttaaagctctgggaggctgaatcctttatttcgctggaagaaaaactgaatgccaagagggcctcactttccccaaagctatacagccatcaggggcaatgctgggattcccccatggatctcttgactcctaatacagtgctctttctgatatgccatctgGCTCCATAATGAACATTGTGTTCCAGGAAAATCAGTCTGCGCTGAACAGGATGGAATTGGGGCAGGACGTCCAGTGAGGAGGACATTGAGGCAGTAGAGCAGTTGATTGCCGAATGACCACCTACCCTGACTTAAAAGATCAACCTCAGGGAGGATTGGAGCTTTCTAGAGTCTCTTGGGACAGCAGAAGCACAGGCCGGGTTGGACTGAATCTTTAAGTGGACATGAGGGACAAAGTACCTCCTGTTGGTAAACATCTCACCACCAACCCACTGTGGTGGCTAAAATCCCACCTTTAGTCCCAGCAACATATGAGCATGTCACCAGGAGGTCTTCACAGGCCTGTCTGCCACCTGAGTGTAGACATCTTTTGGCCCTGGAGCCCAGAGAGGCTGAATGTGGAGAGGGTGGAGAGAGGTCTGTAGTCCTCAGAGAAGACTTGCAGCTTTTTCAGAGCCACCAACCCATAAAAAAAAAAATCCCCAAACAGAAAAATCCTAACTGTGGTGACCAGGTACCTCCTGAGACATGAAGCCTCCACTTACCTGGACCCTGGAGGCCTCTCCGGGCACAGCTGCCAATGTTGATCTTAGATAAGACCACCAGCAAGTAAACATCCACTGTGCAAAGCTGTGTTATCTTTGGGGAACTGAAATGTCCCCTGGGAGTTGGAAACTCCCCTAGCCACATACCACAGAGTTGAGGAAGGAAGAGCTGATGGACGGAAGAACCATGGCGGGAGGAGTCATCCGGAAGCTACCTCGCTGCCCTGTCAGTTACGGAACAGAGGAGAGATGCCGGCTGGAGGACACAGCAAATTTGAACCAAGAGGAGCTTGGAGGAAGCCCGAGCGACCTGGAGGGGACTGGCTGACCTTCCTCATTCTTTTCAAGTGTGAATAATAACCAAGCCCAGTTTGGCAACTCCTTGAGGGTGAGGACGAAGCCCCATTCTCCTTTTTGGAACTTGGTGGGGCTCAGGAAGCAGGTTCTCTCCAGTCGGTGGCTTTCCTTTCTGTTGCGGGTCTCTTGAGGGCCTGCCTTCATGAAGGCACATGAGTGACTCATCATTTGTGAATTAATTGCTATATGTGAAGGGCATCTGAGAACAAATTATCTTCATAGACTTTTCATTATAATTTTATTTGTACTGA</dnaseq>| | sequence = <dnaseq>ACATGGAAGTCTCCAAGCCTGTGCCATCCACCTGCCAAGGAAAAGCACAAGCATACAGTTTGAACATTTATTCGCCTCCTGAGCTTGTACAACAGTCGTGGGAGTTGCTGCAGAAGCAAGCGAAAAGCCAGATGAGCGCTTCTAAACTTAGAGAGAGGGAGAGCGCCTCATCTCTTCCATTTTCCAGGTATTCCTGAGATGATTTATTGGAGCTCAAAGCTTTGGGAGAGTTGGGGCCTTCCATTCCCTCCAGTAAATACTTGTTTTTCTTCCACCGCTGAGGCAAATGCGGGGTGGCTGATCACCTGGCAGACATCTTAGGAAACAGGAGCACCGGTCTGGGAAACTGCTGGCCTGGCCTAACACCTGGCGCTGTGGTGCAGGAGGGCAGCCACTGAGGATTCTTGAGGAGAGAGAGaccatagaatgaatgaaagagttggaaagactttaaagctctgggaggctgaatcctttatttcgctggaagaaaaactgaatgccaagagggcctcactttccccaaagctatacagccatcaggggcaatgctgggattcccccatggatctcttgactcctaatacagtgctctttctgatatgccatctgGCTCCATAATGAACATTGTGTTCCAGGAAAATCAGTCTGCGCTGAACAGGATGGAATTGGGGCAGGACGTCCAGTGAGGAGGACATTGAGGCAGTAGAGCAGTTGATTGCCGAATGACCACCTACCCTGACTTAAAAGATCAACCTCAGGGAGGATTGGAGCTTTCTAGAGTCTCTTGGGACAGCAGAAGCACAGGCCGGGTTGGACTGAATCTTTAAGTGGACATGAGGGACAAAGTACCTCCTGTTGGTAAACATCTCACCACCAACCCACTGTGGTGGCTAAAATCCCACCTTTAGTCCCAGCAACATATGAGCATGTCACCAGGAGGTCTTCACAGGCCTGTCTGCCACCTGAGTGTAGACATCTTTTGGCCCTGGAGCCCAGAGAGGCTGAATGTGGAGAGGGTGGAGAGAGGTCTGTAGTCCTCAGAGAAGACTTGCAGCTTTTTCAGAGCCACCAACCCATAAAAAAAAAAATCCCCAAACAGAAAAATCCTAACTGTGGTGACCAGGTACCTCCTGAGACATGAAGCCTCCACTTACCTGGACCCTGGAGGCCTCTCCGGGCACAGCTGCCAATGTTGATCTTAGATAAGACCACCAGCAAGTAAACATCCACTGTGCAAAGCTGTGTTATCTTTGGGGAACTGAAATGTCCCCTGGGAGTTGGAAACTCCCCTAGCCACATACCACAGAGTTGAGGAAGGAAGAGCTGATGGACGGAAGAACCATGGCGGGAGGAGTCATCCGGAAGCTACCTCGCTGCCCTGTCAGTTACGGAACAGAGGAGAGATGCCGGCTGGAGGACACAGCAAATTTGAACCAAGAGGAGCTTGGAGGAAGCCCGAGCGACCTGGAGGGGACTGGCTGACCTTCCTCATTCTTTTCAAGTGTGAATAATAACCAAGCCCAGTTTGGCAACTCCTTGAGGGTGAGGACGAAGCCCCATTCTCCTTTTTGGAACTTGGTGGGGCTCAGGAAGCAGGTTCTCTCCAGTCGGTGGCTTTCCTTTCTGTTGCGGGTCTCTTGAGGGCCTGCCTTCATGAAGGCACATGAGTGACTCATCATTTGTGAATTAATTGCTATATGTGAAGGGCATCTGAGAACAAATTATCTTCATAGACTTTTCATTATAATTTTATTTGTACTGA</dnaseq>| | ||
}} | }} | ||
| − | [[Category: | + | [[Category:Antisense]][[Category:NONHSAG034421]][[Category:Transcripts]] |
Latest revision as of 06:25, 25 June 2016
GHRLOS, which gives rise to endogenous ghrelin natural antisense transcripts. exhibits features which are common to many non-coding RNA genes, including extensive splicing, lack of significant and conserved open reading frames, differential expression and lack of conservation in vertebrates.
Contents
Annotated Information
Name
GHRLOS:ghrelin opposite strand/antisense RNA(HGNC nomenclature)
"GHRL antisense RNA 1 (non-protein coding)", GHRL-AS1, NCRNA00068, "non-protein coding RNA 68"[1]
Characteristics
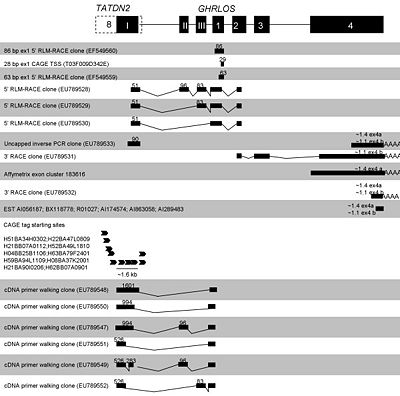
GHRLOS transcripts that extend over 1.4 kb of the promoter region and 106 nucleotides of exon 4 of the ghrelin gene(GHRL) initiate 4.8 kb downstream of the terminal exon 4 of GHRL and are present in the 3' untranslated exon of the adjacent gene TATDN2 (TatD DNase domain containing 2). The 3' region of GHRLOS is also antisense, in a tail-to-tail fashion to a novel terminal exon of the neighbouring SEC13 gene, which is important inprotein transport. GHRLOS that is riddled with stop codons is little nucleotide and amino-acid sequence conservation between vertebrates. The gene spans 44 kb on 3p25.3, is extensively spliced and harbours multiple variable exons[1].
Function
GHRLOS RNA transcripts display several distinctive features of non-coding (ncRNA) genes, including 5' capping, polyadenylation, extensive splicing and short open reading frames[1]. GHRLOS natural antisense transcripts, functioning as a non-coding RNA,may regulate one or both of these genes, because GHRLOS overlaps the terminal exons of both GHRL andSEC13-T[1]
Expression
We have also investigated the expression of GHRLOS and found evidence of differential tissue expression. It is highly expressed in tissues which are emerging as major sites of non-coding RNA expression (the thymus, brain, and testis), as well as in the ovary and uterus. In contrast, very low levels were found in the stomach where sense, GHRL derived RNAs are highly expressed. For non-quantitative RT-PCR analysis of GHRLOS splicing, RT-PCRs were performed with a forward primer in a region common to the 5' terminal exon Ia/b and a reverse primer in the 3' terminal exon 4 of GHRLOS[1].
| Ito4-F | CATGGAAGTCTCCAAGCCTG[1] |
| Ito4-R | CTGCTCTACTGCCTCAATGTC[1] |
To detect long, chimaeric transcripts, we employed RT-PCR with a forward primer in exon 2 of TATDN2(ChiOut-F) and a reverse primer in exon 1 of GHRLOS[1].
| ChiOut-F | TGAAAGCCCAGAAGGAGGA[1] |
| ChiOut-R | TCTAAGTTTAGAAGCGCTCATCTG[1] |
To allow strand-specific and RNA-specific amplification of GHRLOS transcripts, reverse transcription was performed using a gene-specific primer in exon 4 with a linker (LK) sequence attached to the 5' end of the primer[1].
| GHRLOS-Real-F | CATTGAGGCAGTAGAGCAGTTGA[1] |
| LK | CGACTGGAGCACGAGGACACTGA[1] |
To detect sense GHRL transcripts,a strand-specific RT-PCR approach was employed, with a reverse transcription primer spanning the 3' terminal exon 4 of the ghrelin gene (GHRLex4_RT_LK) followed by PCR with an exon 4 specific forward primer (GHRLex4_F) and a linker-specific reverse primer(LK)[1].
| GHRL-Real-RT-LK | CGACTGGAGCACGAGGACACTGAGCCAGAGAGCGCTTCTAAACTTA[1] |
| GHRL-Real-F | GCCCCAGCCGACAAGTG[1] |
| LK | CGACTGGAGCACGAGGACACTGA[1] |
Labs working on this lncRNA
Institute of Health and Biomedical Innovation, Queensland University of Technology, Kelvin Grove, Queensland, Australia[1].
References
Basic Information
| Transcript ID |
NONHSAT087920 |
| Source |
NONCODE4.0 |
| Same with |
GHRLOS, GHRL-AS1, NCRNA00068 |
| Classification |
antisense |
| Length |
1765 nt |
| Genomic location |
chr3+:10322636..10335133 |
| Exon number |
5 |
| Exons |
10322636..10322686,10327432..10327537,10329098..10329165,10329629..10329786,10333752..10335133 |
| Genome context |
|
| Sequence |
000001 ACATGGAAGT CTCCAAGCCT GTGCCATCCA CCTGCCAAGG AAAAGCACAA GCATACAGTT TGAACATTTA TTCGCCTCCT 000080 000081 GAGCTTGTAC AACAGTCGTG GGAGTTGCTG CAGAAGCAAG CGAAAAGCCA GATGAGCGCT TCTAAACTTA GAGAGAGGGA 000160 000161 GAGCGCCTCA TCTCTTCCAT TTTCCAGGTA TTCCTGAGAT GATTTATTGG AGCTCAAAGC TTTGGGAGAG TTGGGGCCTT 000240 000241 CCATTCCCTC CAGTAAATAC TTGTTTTTCT TCCACCGCTG AGGCAAATGC GGGGTGGCTG ATCACCTGGC AGACATCTTA 000320 000321 GGAAACAGGA GCACCGGTCT GGGAAACTGC TGGCCTGGCC TAACACCTGG CGCTGTGGTG CAGGAGGGCA GCCACTGAGG 000400 000401 ATTCTTGAGG AGAGAGAGac catagaatga atgaaagagt tggaaagact ttaaagctct gggaggctga atcctttatt 000480 000481 tcgctggaag aaaaactgaa tgccaagagg gcctcacttt ccccaaagct atacagccat caggggcaat gctgggattc 000560 000561 ccccatggat ctcttgactc ctaatacagt gctctttctg atatgccatc tgGCTCCATA ATGAACATTG TGTTCCAGGA 000640 000641 AAATCAGTCT GCGCTGAACA GGATGGAATT GGGGCAGGAC GTCCAGTGAG GAGGACATTG AGGCAGTAGA GCAGTTGATT 000720 000721 GCCGAATGAC CACCTACCCT GACTTAAAAG ATCAACCTCA GGGAGGATTG GAGCTTTCTA GAGTCTCTTG GGACAGCAGA 000800 000801 AGCACAGGCC GGGTTGGACT GAATCTTTAA GTGGACATGA GGGACAAAGT ACCTCCTGTT GGTAAACATC TCACCACCAA 000880 000881 CCCACTGTGG TGGCTAAAAT CCCACCTTTA GTCCCAGCAA CATATGAGCA TGTCACCAGG AGGTCTTCAC AGGCCTGTCT 000960 000961 GCCACCTGAG TGTAGACATC TTTTGGCCCT GGAGCCCAGA GAGGCTGAAT GTGGAGAGGG TGGAGAGAGG TCTGTAGTCC 001040 001041 TCAGAGAAGA CTTGCAGCTT TTTCAGAGCC ACCAACCCAT AAAAAAAAAA ATCCCCAAAC AGAAAAATCC TAACTGTGGT 001120 001121 GACCAGGTAC CTCCTGAGAC ATGAAGCCTC CACTTACCTG GACCCTGGAG GCCTCTCCGG GCACAGCTGC CAATGTTG |