Difference between revisions of "NONHSAT099821"
Chunlei Yu (talk | contribs) |
Chunlei Yu (talk | contribs) |
||
| Line 60: | Line 60: | ||
tID = NONHSAT099821| | tID = NONHSAT099821| | ||
source = NONCODE4.0| | source = NONCODE4.0| | ||
| − | same =''DBE-T'' ,"chromatin-associated long non-coding RNA", | + | same =''DBE-T'' ,"chromatin-associated long non-coding RNA", DBET, DUX4L30| |
classification = intergenic| | classification = intergenic| | ||
length = 682 nt| | length = 682 nt| | ||
Revision as of 04:45, 4 April 2015
DBE-T, a chromatin-associated noncoding RNA expressed selectively in FSHD patients, directly recruits ASH1L to the FSHD locus to co-ordinate 4q35 gene de-repression[1]
Contents
Annotated Information
Transcriptomic Nomeclature
DBE-T:D4Z4 binding element transcript(HGNC nomenclature)
"chromatin-associated long non-coding RNA", DBE-T, DUX4L30[1]
Cellular Localization
nuclear[1].
Function
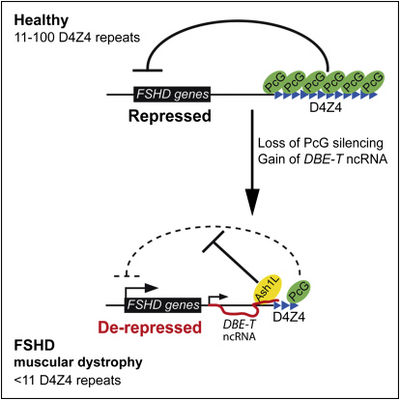
DBE-T recruits the Trithorax group protein Ash1L to the FSHD locus, driving histone H3 lysine 36 dimethylation, chromatin remodeling, and 4q35 gene transcription[1].
Regulation
Ash1L promotes the expression of DBE-T, implying a positive feedback loop that would sustain 4q35 gene de-repression[1].
Diseases
- Facioscapulohumeral muscular dystrophy (FSHD)[1]
Expression
DBE-T knockdown prevented 4q35 gene de-repression caused by AZA plus TSA, strongly suggesting a functional role for this transcript in the positive regulation of 4q35 gene expression[1]
| shRNA | Primer |
|---|---|
| DBE | GCTCACCGCCATTCATGAAGG[1] |
| NDE | GCTCACCGCCATTCATGAAGG[1] |
| Nonsilencing | TCTCGCTTGGGCGAGAGTAAG[1] |
| Ash1L | GCTGGTCATTTATTGCTCAAT[1] |
| Suz12 | TTCTACAAACAGCATACAG[1] |
Labs working on this lncRNA
Dulbecco Telethon Institute at San Raffaele Scientific Institute, Division of Regenerative Medicine, Stem Cells, and Gene Therapy, 20132 Milan, Italy
References
Basic Information
| Transcript ID |
NONHSAT099821 |
| Source |
NONCODE4.0 |
| Same with |
DBE-T ,"chromatin-associated long non-coding RNA", DBET, DUX4L30 |
| Classification |
intergenic |
| Length |
682 nt |
| Genomic location |
chr4+:190986388..190987069 |
| Exon number |
1 |
| Exons |
190986388..190987069 |
| Genome context |
|
| Sequence |
000001 GCCTGGGGCT CTCCCACAGG GGGCTTTCGT GAGCCAGGCA GCGAGGGCCG CCCCCGCGCT GCAGCCCAGC CAGGCCGCGC 000080
000081 CGGCAGAGGG GATCTCCCAA CCTGCCCCGG CGCGCGGGGA TTTCGCCTAC GCCGCCCCGG CTCCTCCGGA GCCGGGGCGC 000160 000161 TCTCCCACCC TCAGGCTCCT CGGTGGCCTC CGCACCCGGG CAAAAGCCGG GAGGACCGGG ACCCGCAGCG CGACGGCCTG 000240 000241 CCGGGCCCCT GCGCGGTGGC ACAGCCTGGG CCCGCTCAAG CGGGGCCGCA GGGCCAAGGG GTGCTTGCGC CACCCACGTC 000320 000321 CCAGGGGAGT CCGTGGTGGG GCTGGGGCCG GGGTCCCCAG GTCGCCGGGG CGGCGTGGGA ACCCCAAGCC GGGGCAGCTC 000400 000401 CACCTCCCCA GCCCGCGCCC CCCGGACGCC TCCGCCTCCG CGCGGCAGGG GCAGATGCAA GGCATCCCGG CGCCCTCCCA 000480 000481 GGCGCTCCAG GAGCCGGCGC CCTGGTCTGC ACTCCCCTGC GGCCTGCTGC TGGATGAGCT CCTGGCGAGC CCGGAGTTTC 000560 000561 TGCAGCAGGC GCAACCTCTC CTAGAAACGG AGGCCCCGGG GGAGCTGGAG GCCTCGGAAG AGGCGCCTCG CTGGAAGCAC 000640 000641 CCCTCAGCGA GGAAGAATAC CGGGCTCTGC TGGAGGAGCT TT |