Difference between revisions of "LINC00850"
Chunlei Yu (talk | contribs) (→Expression) |
Chunlei Yu (talk | contribs) (→Expression) |
||
| Line 37: | Line 37: | ||
|- | |- | ||
| |Dystrophin cDNA | | |Dystrophin cDNA | ||
| − | | | + | | ! |
|- | |- | ||
| rowspan="1"|Exon 18r | | rowspan="1"|Exon 18r | ||
Revision as of 02:51, 11 April 2015
LINC00850 spanning nearly 50kb on Xq28 specially expressed in brain, which indicated a potential role in mental retardation.
Contents
Annotated Information
Name
LINC00850:long intergenic non-protein coding RNA 850(HGNC nomenclature)
KUCG1[1]
Characteristics
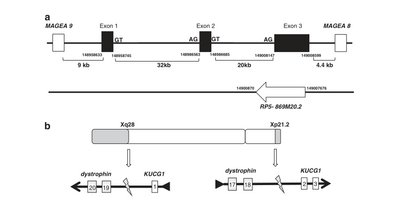
LINC00850(alse named KUCG1) spans nearly 50kb onXq28 and is located 9.0kb downstream of MAGEA1 and 4.4kb upstream of MAGEA8. It has three exons separated by two introns(32kb and 20kb long, respectively). Its intron 1 encompassed the site of recombination of the intrachromosomal inversion inv(X)(p21.2; q28), which cause a a head-to-tail fusion of KUCG1 and dystrophin at the recombination sites. Exon 3 of KUCG1 overlaps with antisense transcript RP5-869M20.2, an lncRNA of unknown function.[1]
It belongs to the category of "Intergenic" in lncRNA classification.
Function
KUCG1 disruption may responsible for the moderate mental retardation in the index case as it was expressed in the brain.[1].
Diseases
- mental retardation
Expression
KUCG1 was specially expressed in brain.[1]
Primer used as follows
| Primer name | Sequence(5'-3') |
|---|---|
| Dystrophin cDNA | ! |
| Exon 18r | GCAGAGTCCTGAATTTGCAATC[1] |
| Exon 19f | CATTCACCATCTGTTCCACCA[1] |
| 5'-RACE | |
| c24r | CAGCCATCCATTTCTTCAGG[1] |
| c21r | TTGTCTGTAGCTCTTTCTCT[1] |
| c20r | ACTGGCAGAATTCGATCCAC[1] |
| 3'-RACE | |
| c16f | CTGATCTAGAGGTACCGGATCC[1] |
| c18f | GCAGAGTCCTGAATTTGCAATC[1] |
| KUCG1 mRNA | |
| Bf | GGTGAACCCCTCAATGTAAG[1] |
| Cr | CTCTTGTATTCGCTGCAGTG[1] |
| Cr2 | CAGCAAACTTGTACAGTTGC[1] |
Sequence
>gi|565324244|ref|NR_109813.1| Homo sapiens long intergenic non-protein coding RNA 850 (LINC00850), long non-coding RNA
000081 AAAATATTTC CTTGGTGGTC AGGACCAAGG CTGGAGGTGA ACCCCTCAAT GTAAGTGTCA AGGGACAACA TTCCTGCCTG 000160
000161 ATATGTGAGC CAATCAGGAG TTCACTACAA TCCGTGAAGC CATAACCAGG GACATTCAAA CTGCAAACCA GGCTGCAGTG 000240
000241 AATGCTGAAA TGAAAAAGAG AGCTATGGAT ATCAAGATGA AAAAGAGGTA TGAAGATTGA AAAGAAATAG ATCATATTTT 000320
000321 TCACAATATG ACCATCAAGA AAATTCAAAG GAAGAGACAC ATGGAACATT CAAATGAATA AGAGACTTCA GAAGACAGCA 000400
000401 TACAAAGACC ACTGCAGCGA ATACAAGAGA CTTGTCACAG ATGAGCAACT TAGAACATAA AATGAAAAAT ATACCATCCC 000480
000481 CAACAGCATT TAAATCCAAA ATATAAAGAA GAAGAAGCCT AACCAAAATA TGCAAGACAT TTACAGAAGC AACTGTACAA 000560
000561 GTTTGCTGAC AGATTCAGAG AAAAAATTCC CAAATAAATG GAGTAGTATA TGATGTTCAC TGATGAGGAA TAGAATACAG 000640
000641 CAAAGGTGTA AATCTTACTT AAATTAAATA AACAGTTAAT GCCTA
Labs working on this lncRNA
- Department of Pediatrics, Kobe University Graduate School of Medicine, Kobe, Japan.[1].
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 Tran TH, Zhang Z, Yagi M, Lee T, Awano H, Nishida A, et al. Molecular characterization of an X(p21.2;q28) chromosomal inversion in a Duchenne muscular dystrophy patient with mental retardation reveals a novel long non-coding gene on Xq28[J]. Journal of human genetics. 2013,58(1):33-9.