Difference between revisions of "SMILR"
| Line 1: | Line 1: | ||
==Annotated Information== | ==Annotated Information== | ||
===Approved Symbol=== | ===Approved Symbol=== | ||
| − | SMILR | + | SMILR: smooth muscle induced lncRNA, enhancer of proliferation <ref name="ref1" /> |
| − | + | RP11-94a24.1 | |
| − | |||
===Chromosome=== | ===Chromosome=== | ||
8q24.13 | 8q24.13 | ||
| Line 9: | Line 8: | ||
NR_131202 | NR_131202 | ||
===Characteristics=== | ===Characteristics=== | ||
| − | The longest open reading frame(ORF) within SMILR is 57 amino acids. | + | SMILR is located ~750 kbp downstream of HAS2 on the same (reverse) strand and ~350 kbp from ZHX2 and ~750 kbp from HAS2-AS1 on the opposite strand of chromosome 8 <ref name="ref1" />. The longest open reading frame(ORF) within SMILR is 57 amino acids <ref name="ref1" />. |
[[File:Localization of SMILR.jpg|right|thumb|300px|'''Localization of SMILR.(A)RNA FISH analysis of SMILR,cytoplasmic UBC mRNA,and nuclear SNORD3 RNA in HSVSMC.(B)25±5 individual SMILR molecules were observed within the nucleus and cytoplasm.''' <ref name="ref1" />.]] | [[File:Localization of SMILR.jpg|right|thumb|300px|'''Localization of SMILR.(A)RNA FISH analysis of SMILR,cytoplasmic UBC mRNA,and nuclear SNORD3 RNA in HSVSMC.(B)25±5 individual SMILR molecules were observed within the nucleus and cytoplasm.''' <ref name="ref1" />.]] | ||
| + | |||
| + | ===Cellular Localization=== | ||
| + | SMILR is located in both nucleus and cytoplasm <ref name="ref1" />. | ||
===Expression=== | ===Expression=== | ||
| − | SMILR is expressed highest in | + | SMILR is expressed highest in heart <ref name="ref1" />. It is differentially expressed in human saphenous vein-derived smooth muscle cells following stimulation with interleukin-1α and platelet-derived growth factor <ref name="ref1" />. Increased expression of SMILR is observed in unstable atherosclerotic plaques, and plasma from patients with high plasma C-reactive protein <ref name="ref1" />. |
| − | |||
| − | |||
| − | SMILR | ||
===Function=== | ===Function=== | ||
| − | SMILR | + | SMILR acts as a driver of vascular smooth muscle cell proliferation <ref name="ref1" />. Knockdown of SMILR markedly reduced cell proliferation and HAS2 expression was also reduced by SMILR knockdown <ref name="ref1" />. |
===Regulation=== | ===Regulation=== | ||
SMILR expression can be regulated by IL1α or PDGF and it is a time-dependent manner <ref name="ref1" />. | SMILR expression can be regulated by IL1α or PDGF and it is a time-dependent manner <ref name="ref1" />. | ||
| − | |||
| − | |||
| − | |||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
| − | * | + | * BHF Glasgow Cardiovascular Research Centre, University of Glasgow, United Kingdom |
| − | + | * British Heart Foundation/University of Edinburgh Centre for Cardiovascular Science, Edinburgh, United Kingdom | |
| − | * | ||
==References== | ==References== | ||
Revision as of 09:12, 25 September 2017
Contents
Annotated Information
Approved Symbol
SMILR: smooth muscle induced lncRNA, enhancer of proliferation [1] RP11-94a24.1
Chromosome
8q24.13
RefSeq(supplied by NCBI)
NR_131202
Characteristics
SMILR is located ~750 kbp downstream of HAS2 on the same (reverse) strand and ~350 kbp from ZHX2 and ~750 kbp from HAS2-AS1 on the opposite strand of chromosome 8 [1]. The longest open reading frame(ORF) within SMILR is 57 amino acids [1].
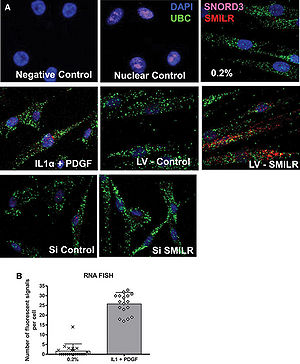
Cellular Localization
SMILR is located in both nucleus and cytoplasm [1].
Expression
SMILR is expressed highest in heart [1]. It is differentially expressed in human saphenous vein-derived smooth muscle cells following stimulation with interleukin-1α and platelet-derived growth factor [1]. Increased expression of SMILR is observed in unstable atherosclerotic plaques, and plasma from patients with high plasma C-reactive protein [1].
Function
SMILR acts as a driver of vascular smooth muscle cell proliferation [1]. Knockdown of SMILR markedly reduced cell proliferation and HAS2 expression was also reduced by SMILR knockdown [1].
Regulation
SMILR expression can be regulated by IL1α or PDGF and it is a time-dependent manner [1].
Labs working on this lncRNA
- BHF Glasgow Cardiovascular Research Centre, University of Glasgow, United Kingdom
- British Heart Foundation/University of Edinburgh Centre for Cardiovascular Science, Edinburgh, United Kingdom