Difference between revisions of "ENST00000534336.1"
(→Expression) |
(→Diseases) |
||
| Line 69: | Line 69: | ||
===Diseases=== | ===Diseases=== | ||
| − | MALAT-1 was first identified as a prognostic marker for metastasis and patient survival in non-small cell lung cancer (NSCLC) <ref name="ref28" />. It is found to be overexpressed in various tumors and cancer cell lines, including | + | MALAT-1 was first identified as a prognostic marker for metastasis and patient survival in non-small cell lung cancer (NSCLC) <ref name="ref28" />. It is found to be overexpressed in various tumors and cancer cell lines, including lung cancer <ref name="ref1" /><ref name="ref21" /><ref name="ref28" />, endometrial stromal sarcoma of the uterus <ref name="ref29" />, hepatocellular carcinomas <ref name="ref30" /><ref name="ref31" />, breast cancer <ref name="ref21" /><ref name="ref32" />, pancreas cancer <ref name="ref21" />, colon cancer <ref name="ref21" />, prostate cancer <ref name="ref15" />, melanoma <ref name="ref22" />, bladder cancer <ref name="ref18" />. Overexpression of MALAT-1 in cancer cells is closely associated with tumor growth and metastasis <ref name="ref12" /><ref name="ref20" /><ref name="ref22" /><ref name="ref28" />. |
There is no significant difference in MALAT-1 lncRNA levels in normal pituitary tissues, invasive NFPAs (non-functioning pituitary adenomas), and non-invasive NFPAs, and no significant association between MALAT-1 expression and patient clinicopathological characteristics <ref name="ref23" />. | There is no significant difference in MALAT-1 lncRNA levels in normal pituitary tissues, invasive NFPAs (non-functioning pituitary adenomas), and non-invasive NFPAs, and no significant association between MALAT-1 expression and patient clinicopathological characteristics <ref name="ref23" />. | ||
Revision as of 08:14, 29 June 2014
Please input one-sentence summary here.
Contents
Annotated Information
Transcriptomic Nomeclature
hsa-N11QT0096001-SSCY-LMXXX01 help
Name
Malat1: Metastasis-associated lung adenocarcinoma transcript 1
Neat2: Nuclear enriched abundant transcript 2
Characteristics
Malat1 gene is localized in the intergenic region of the genome. It is about 8kb in length and the transcript has only one exon [1][2]. The 3’-end of the transcript is a conserved tRNA-like sequence, which can be modified by RNase P and cleaved by RNase Z to yield another ncRNA (61nt), the cytoplasmic MALAT1-associated small cytoplasmic RNA (mascRNA) [3].
Malat1 is stable in human B cells (half-life: 16.5 h) [4] and Hela cells (half-life: about 7 h) [5], but is unstable in mouse 3T3 cells (half-life: 3 h) [4] and N2A cells ( half-life: 4 h) [4]. MALAT-1 is generally stable in cancer cells, with the half-life ranging from ~ 9 h to > 12 h in various cancer cells [6]. Xrn2, PM/Scl-75, PARN, and Mtr4, known nuclear RNases or RNA helicases, do not affect MALAT-1 degradation [6].
Cellular localization
MALAT-1 localizes predominately in the nucleus [2]. In G2/M cell cycle phase, MALAT-1 transcripts partially translocate from the nucleus into the cytoplasm [7]. RNPS1, SRm160, and IBP160 are found to contribute to the nuclear localization of MALAT-1 [8].
Function
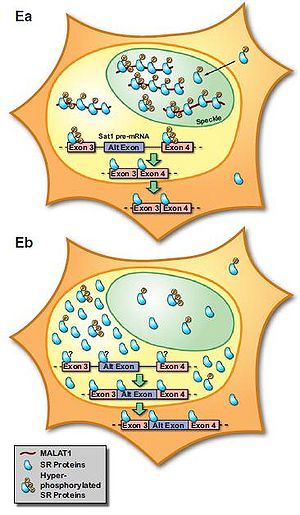
MALAT-1 localizes specifically in the SC35 splicing domains in the nucleus, suggesting its function in pre-mRNA metabolism or specific nuclear structures [2]. The later studies found that MALAT-1 could modulate mRNA alternative splicing via its interaction with the serine/arginine-rich (SR) family of nuclear phosphoproteins that are involved in the splicing machinery [9]. MALAT-1 regulates synapse formation by modulating the expression of genes involved in synapse formation and/or maintenance [10]. However, splicing alterations were not found after Malat1 ablation in mice [11]. Also, MALAT1 does not alter alternative splicing but actively regulates gene expression including a set of metastasis-associated genes in lung cancer cells [12]. These results indicate that alternative functions for MALAT-1 may exist.
MALAT-1 can function by participating in localization of important proteins, such as hnRNP C [7] and growth control genes [13]. MALAT-1 is found to regulate cell cycle progress in G2/M phase [7][9], G1/S phase [14], and G0/G1 phase [15]. In the G2/M phase, MALAT-1 interacts with hnRNP C to facilitate the cytoplasmic translocation of hnRNP C, leading to cell cycle progresion [7]. MALAT1 could interact with the demethylated form of CBX4 (chromobox homolog 4), and controls the relocalization of growth control genes between polycomb bodies and interchromatin granules [13]. MALAT1 could bind to Human PSF (hPSF) protein to release hPSF from a repressed proto-oncogene and activate transcription, driving transformation and tumorigenesis [16].
MALAT-1 regulates synaptogenesis [10]and is involved in the development of advanced invasive placentation [17]. Deregulation of MALAT-1 is found to be closely associated with the development of cancer. In vitro, it is found that MALAT-1 promotes epithelial–mesenchymal transition (EMT) of bladder cancer cells by activating Wnt signaling [18]. 3' end of MALAT-1 (6918 nt-8441 nt) is found to be important in colorectal cancer metastasis [19]. In lung adenocarcinoma cells, MALAT-1 may regulate cell motility through transcriptional and post-transcriptional regulation of motility related gene expression [20]. However, mechanisms of these functions are not clear.
Expression
MALAT-1 is ubiquity expressed in various normal tissues [1][2][3][10], but the expression levels are quite different among tissues [1][2][10].
MALAT-1 is over-expressed in many human carcinomas, including those of the breast, pancreas, lung, colon, prostate, and liver [21].
It is also found to be up-regulated in the cerebellum, hippocampus and brain stem of human alcoholics [22].
| Primer | Forward primer | Reverse primer |
|---|---|---|
| RT-PCR | 5'-AAAGCAAGGTCTCCCCACAAG-3' | 5'-GGTCTGTGCTAGATCAAAAGGCA-3'[23][24] |
| 5'-CTTCCCTAGGGGATTTCAGG-3' | 5'-GCCCACAGGAACAAGTCCTA-3'[15] | |
| 5'-GAATTGCGTCATTTAAAGCCTAGTT-3' | 5'-GTTTCATCCTACCACTCCCAATTAAT-3'[25] | |
| 5'-cggaagtaattcaagatcaagag-3' | 5'-actgaatccacttctgtgtagc-3'[16] | |
| cDNA amplication | 5'-GTAGGGCCCTCCATGGCGATTTGCCTTGTGAGCAC-3' | 5'-GAGCTCGAGGTCCTGAAGACAGATTAGTAGTCAAAGC-3'[6] |
| Northern blot | 5'-GGCAGGAGAGACAACAAAGC-3' | 5'-CTCGACACCATCGTTACCT-3'[2] |
Regulation
In breast cancer cells, high concentration E2 treatment largely decreases MALAT-1 RNA level in an ERa independent way [26].
Disruption of p53 appears to play an important role in the up-regulation of MALAT-1 [27].
CREB (cyclic AMP-responsive element binding) transcription factor is found to bind to the defined proximal promoter of the MALAT1 gene, leading to the up-regulation of MALAT1 [28].
Diseases
MALAT-1 was first identified as a prognostic marker for metastasis and patient survival in non-small cell lung cancer (NSCLC) [29]. It is found to be overexpressed in various tumors and cancer cell lines, including lung cancer [1][22][29], endometrial stromal sarcoma of the uterus [30], hepatocellular carcinomas [21][31], breast cancer [22][32], pancreas cancer [22], colon cancer [22], prostate cancer [15], melanoma [23], bladder cancer [18]. Overexpression of MALAT-1 in cancer cells is closely associated with tumor growth and metastasis [12][20][23][29].
There is no significant difference in MALAT-1 lncRNA levels in normal pituitary tissues, invasive NFPAs (non-functioning pituitary adenomas), and non-invasive NFPAs, and no significant association between MALAT-1 expression and patient clinicopathological characteristics [24].
Evolution
MALAT-1 is highly conserved over its full length (~8 kb) across mammals [10]. However, sequence conservation is limited in vertebrates. Sequence similarity between zebrafish and mammalian MALAT1 is restricted to the 3′ end, while the length of MALAT1 (~7 kb) along with the gene structure appeare to be roughly fixed in all vertebrates [14].
Associated components
hnRNP C [7]
CBX4 (chromobox homolog 4) [13]
Human PSF (hPSF) protein [16]
You can also add sub-section(s) at will.
Labs working on this lncRNA
Please input related labs here.
References
- ↑ 1.0 1.1 1.2 1.3 Ji, P., Diederichs, S., Wang, W., Boing, S., Metzger, R., Schneider, P.M., Tidow, N., Brandt, B., Buerger, H., Bulk, E. et al. (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene, 22, 8031-8041.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Hutchinson, J.N., Ensminger, A.W., Clemson, C.M., Lynch, C.R., Lawrence, J.B. and Chess, A. (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics, 8, 39.
- ↑ 3.0 3.1 Wilusz, J.E., Freier, S.M. and Spector, D.L. (2008) 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell, 135, 919-932.
- ↑ 4.0 4.1 4.2 Clark, M.B., Johnston, R.L., Inostroza-Ponta, M., Fox, A.H., Fortini, E., Moscato, P., Dinger, M.E. and Mattick, J.S. (2012) Genome-wide analysis of long noncoding RNA stability. Genome Res, 22, 885-898.
- ↑ Tani, H., Mizutani, R., Salam, K.A., Tano, K., Ijiri, K., Wakamatsu, A., Isogai, T., Suzuki, Y. and Akimitsu, N. (2012) Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res, 22, 947-956.
- ↑ 6.0 6.1 6.2 Tani, H., Nakamura, Y., Ijiri, K. and Akimitsu, N. (2010) Stability of MALAT-1, a nuclear long non-coding RNA in mammalian cells, varies in various cancer cells. Drug Discov Ther, 4, 235-239.
- ↑ 7.0 7.1 7.2 7.3 7.4 Yang, F., Yi, F., Han, X., Du, Q. and Liang, Z. (2013) MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett, 587, 3175-3181.
- ↑ Miyagawa, R., Tano, K., Mizuno, R., Nakamura, Y., Ijiri, K., Rakwal, R., Shibato, J., Masuo, Y., Mayeda, A., Hirose, T. et al. (2012) Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA, 18, 738-751.
- ↑ 9.0 9.1 9.2 Tripathi, V., Ellis, J.D., Shen, Z., Song, D.Y., Pan, Q., Watt, A.T., Freier, S.M., Bennett, C.F., Sharma, A., Bubulya, P.A. et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell, 39, 925-938.
- ↑ 10.0 10.1 10.2 10.3 10.4 Bernard, D., Prasanth, K.V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., Zhang, M.Q., Sedel, F., Jourdren, L., Coulpier, F. et al. (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J, 29, 3082-3093.
- ↑ Zhang, B., Arun, G., Mao, Y.S., Lazar, Z., Hung, G., Bhattacharjee, G., Xiao, X., Booth, C.J., Wu, J., Zhang, C. et al. (2012) The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep, 2, 111-123.
- ↑ 12.0 12.1 Gutschner, T., Hammerle, M., Eissmann, M., Hsu, J., Kim, Y., Hung, G., Revenko, A., Arun, G., Stentrup, M., Gross, M. et al. (2013) The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res, 73, 1180-1189.
- ↑ 13.0 13.1 13.2 Yang, L., Lin, C., Liu, W., Zhang, J., Ohgi, K.A., Grinstein, J.D., Dorrestein, P.C. and Rosenfeld, M.G. (2011) ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell, 147, 773-788.
- ↑ 14.0 14.1 Ulitsky, I., Shkumatava, A., Jan, C.H., Sive, H. and Bartel, D.P. (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell, 147, 1537-1550.
- ↑ 15.0 15.1 15.2 Ren, S., Liu, Y., Xu, W., Sun, Y., Lu, J., Wang, F., Wei, M., Shen, J., Hou, J., Gao, X. et al. (2013) Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol, 190, 2278-2287.
- ↑ 16.0 16.1 16.2 Li, L., Feng, T., Lian, Y., Zhang, G., Garen, A. and Song, X. (2009) Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A, 106, 12956-12961.
- ↑ Tseng, J.J., Hsieh, Y.T., Hsu, S.L. and Chou, M.M. (2009) Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod, 15, 725-731.
- ↑ 18.0 18.1 Ying, L., Chen, Q., Wang, Y., Zhou, Z., Huang, Y. and Qiu, F. (2012) Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst, 8, 2289-2294.
- ↑ Xu, C., Yang, M., Tian, J., Wang, X. and Li, Z. (2011) MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol, 39, 169-175.
- ↑ 20.0 20.1 Tano, K., Mizuno, R., Okada, T., Rakwal, R., Shibato, J., Masuo, Y., Ijiri, K. and Akimitsu, N. (2010) MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett, 584, 4575-4580.
- ↑ 21.0 21.1 Lin, R., Maeda, S., Liu, C., Karin, M. and Edgington, T.S. (2007) A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene, 26, 851-858.
- ↑ 22.0 22.1 22.2 22.3 22.4 Kryger, R., Fan, L., Wilce, P.A. and Jaquet, V. (2012) MALAT-1, a non protein-coding RNA is upregulated in the cerebellum, hippocampus and brain stem of human alcoholics. Alcohol, 46, 629-634.
- ↑ 23.0 23.1 23.2 Tian, Y., Zhang, X., Hao, Y., Fang, Z. and He, Y. (2014) Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res.
- ↑ 24.0 24.1 Li, Z., Li, C., Liu, C., Yu, S. and Zhang, Y. (2014) Expression of the long non-coding RNAs MEG3, HOTAIR, and MALAT-1 in non-functioning pituitary adenomas and their relationship to tumor behavior. Pituitary.
- ↑ Guo, F., Li, Y., Liu, Y., Wang, J. and Li, G. (2010) Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai), 42, 224-229.
- ↑ Zhao, Z., Chen, C., Liu, Y. and Wu, C. (2014) 17beta-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem Biophys Res Commun, 445, 388-393.
- ↑ Jeffers, L.K., Duan, K., Ellies, L.G., Seaman, W.T., Burger-Calderon, R.A., Diatchenko, L.B. and Webster-Cyriaque, J. (2013) Correlation of transcription of MALAT-1, a novel noncoding RNA, with deregulated expression of tumor suppressor p53 in small DNA tumor virus models. J Cancer Ther, 4.
- ↑ Koshimizu, T.A., Fujiwara, Y., Sakai, N., Shibata, K. and Tsuchiya, H. (2010) Oxytocin stimulates expression of a noncoding RNA tumor marker in a human neuroblastoma cell line. Life Sci, 86, 455-460.
- ↑ 29.0 29.1 29.2 Schmidt, L.H., Spieker, T., Koschmieder, S., Schaffers, S., Humberg, J., Jungen, D., Bulk, E., Hascher, A., Wittmer, D., Marra, A. et al. (2011) The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol, 6, 1984-1992.
- ↑ Yamada, K., Kano, J., Tsunoda, H., Yoshikawa, H., Okubo, C., Ishiyama, T. and Noguchi, M. (2006) Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci, 97, 106-112.
- ↑ Lai, M.C., Yang, Z., Zhou, L., Zhu, Q.Q., Xie, H.Y., Zhang, F., Wu, L.M., Chen, L.M. and Zheng, S.S. (2011) Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol, 29, 1810-1816.
- ↑ Guffanti, A., Iacono, M., Pelucchi, P., Kim, N., Solda, G., Croft, L.J., Taft, R.J., Rizzi, E., Askarian-Amiri, M., Bonnal, R.J. et al. (2009) A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics, 10, 163.
Basic Information
| Transcript ID |
ENST00000534336.1 |
| Source |
{{{source}}} |
| Same with |
lnc-SCYL1-1:2;n343070;Malat1(NR_002819.2) |
| Classification |
intergenic |
| Length |
{{{length}}} |
| Genomic location |
chr11+:65265233-65273940 |
| Exon number |
{{{number}}} |
| Exons |
{{{exons}}} |
| Genome context |
{{{context}}} |
| Sequence |
000001 GTAAAGGACT GGGGCCCCGC AACTGGCCTC TCCTGCCCTC TTAAGCGCAG CGCCATTTTA GCAACGCAGA AGCCCGGCGC 000080
000081 CGGGAAGCCT CAGCTCGCCT GAAGGCAGGT CCCCTCTGAC GCCTCCGGGA GCCCAGGTTT CCCAGAGTCC TTGGGACGCA 000160 000161 GCGACGAGTT GTGCTGCTAT CTTAGCTGTC CTTATAGGCT GGCCATTCCA GGTGGTGGTA TTTAGATAAA ACCACTCAAA 000240 000241 CTCTGCAGTT TGGTCTTGGG GTTTGGAGGA AAGCTTTTAT TTTTCTTCCT GCTCCGGTTC AGAAGGTCTG AAGCTCATAC 000320 000321 CTAACCAGGC ATAACACAGA ATCTGCAAAA CAAAAACCCC TAAAAAAGCA GACCCAGAGC AGTGTAAACA CTTCTGGGTG 000400 000401 TGTCCCTGAC TGGCTGCCCA AGGTCTCTGT GTCTTCGGAG ACAAAGCCAT TCGCTTAGTT GGTCTACTTT AAAAGGCCAC 000480 000481 TTGAACTCGC TTTCCATGGC GATTTGCCTT GTGAGCACTT TCAGGAGAGC CTGGAAGCTG AAAAACGGTA GAAAAATTTC 000560 000561 CGTGCGGGCC GTGGGGGGCT GGCGGCAACT GGGGGGCCGC AGATCAGAGT GGGCCACTGG CAGCCAACGG CCCCCGGGGC 000640 000641 TCAGGCGGGG AGCAGCTCTG TGGTGTGGGA TTGAGGCGTT TTCCAAGAGT GGGTTTTCAC GTTTCTAAGA TTTCCCAAGC 000720 000721 AGACAGCCCG TGCTGCTCCG ATTTCTCGAA CAAAAAAGCA AAACGTGTGG CTGTCTTGGG AGCAAGTCGC AGGACTGCAA 000800 000801 GCAGTTGGGG GAGAAAGTCC GCCATTTTGC CACTTCTCAA CCGTCCCTGC AAGGCTGGGG CTCAGTTGCG TAATGGAAAG 000880 000881 TAAAGCCCTG AACTATCACA CTTTAATCTT CCTTCAAAAG GTGGTAAACT ATACCTACTG TCCCTCAAGA GAACACAAGA 000960 000961 AGTGCTTTAA GAGGTATTTT AAAAGTTCCG GGGGTTTTGT GAGGTGTTTG ATGACCCGTT TAAAATATGA TTTCCATGTT 001040 001041 TCTTTTGTCT AAAGTTTGCA GCTCAAATCT TTCCACACGC TAGTAATTTA AGTATTTCTG CATGTGTAGT TTGCATTCAA 001120 001121 GTTCCATAAG CTGTTAAGAA AAATCTAGAA AAGTAAAACT AGAACCTATT TTTAACCGAA GAACTACTTT TTGCCTCCCT 001200 001201 CACAAAGGCG GCGGAAGGTG ATCGAATTCC GGTGATGCGA GTTGTTCTCC GTCTATAAAT ACGCCTCGCC CGAGCTGTGC 001280 001281 GGTAGGCATT GAGGCAGCCA GCGCAGGGGC TTCTGCTGAG GGGGCAGGCG GAGCTTGAGG AAACCGCAGA TAAGTTTTTT 001360 001361 TCTCTTTGAA AGATAGAGAT TAATACAACT ACTTAAAAAA TATAGTCAAT AGGTTACTAA GATATTGCTT AGCGTTAAGT 001440 001441 TTTTAACGTA ATTTTAATAG CTTAAGATTT TAAGAGAAAA TATGAAGACT TAGAAGAGTA GCATGAGGAA GGAAAAGATA 001520 001521 AAAGGTTTCT AAAACATGAC GGAGGTTGAG ATGAAGCTTC TTCATGGAGT AAAAAATGTA TTTAAAAGAA AATTGAGAGA 001600 001601 AAGGACTACA GAGCCCCGAA TTAATACCAA TAGAAGGGCA ATGCTTTTAG ATTAAAATGA AGGTGACTTA AACAGCTTAA 001680 001681 AGTTTAGTTT AAAAGTTGTA GGTGATTAAA ATAATTTGAA GGCGATCTTT TAAAAAGAGA TTAAACCGAA GGTGATTAAA 001760 001761 AGACCTTGAA ATCCATGACG CAGGGAGAAT TGCGTCATTT AAAGCCTAGT TAACGCATTT ACTAAACGCA GACGAAAATG 001840 001841 GAAAGATTAA TTGGGAGTGG TAGGATGAAA CAATTTGGAG AAGATAGAAG TTTGAAGTGG AAAACTGGAA GACAGAAGTA 001920 001921 CGGGAAGGCG AAGAAAAGAA TAGAGAAGAT AGGGAAATTA GAAGATAAAA ACATACTTTT AGAAGAAAAA AGATAAATTT 002000 002001 AAACCTGAAA AGTAGGAAGC AGAAGAAAAA AGACAAGCTA GGAAACAAAA AGCTAAGGGC AAAATGTACA AACTTAGAAG 002080 002081 AAAATTGGAA GATAGAAACA AGATAGAAAA TGAAAATATT GTCAAGAGTT TCAGATAGAA AATGAAAAAC AAGCTAAGAC 002160 002161 AAGTATTGGA GAAGTATAGA AGATAGAAAA ATATAAAGCC AAAAATTGGA TAAAATAGCA CTGAAAAAAT GAGGAAATTA 002240 002241 TTGGTAACCA ATTTATTTTA AAAGCCCATC AATTTAATTT CTGGTGGTGC AGAAGTTAGA AGGTAAAGCT TGAGAAGATG 002320 002321 AGGGTGTTTA CGTAGACCAG AACCAATTTA GAAGAATACT TGAAGCTAGA AGGGGAAGTT GGTTAAAAAT CACATCAAAA 002400 002401 AGCTACTAAA AGGACTGGTG TAATTTAAAA AAAACTAAGG CAGAAGGCTT TTGGAAGAGT TAGAAGAATT TGGAAGGCCT 002480 002481 TAAATATAGT AGCTTAGTTT GAAAAATGTG AAGGACTTTC GTAACGGAAG TAATTCAAGA TCAAGAGTAA TTACCAACTT 002560 002561 AATGTTTTTG CATTGGACTT TGAGTTAAGA TTATTTTTTA AATCCTGAGG ACTAGCATTA ATTGACAGCT GACCCAGGTG 002640 002641 CTACACAGAA GTGGATTCAG TGAATCTAGG AAGACAGCAG CAGACAGGAT TCCAGGAACC AGTGTTTGAT GAAGCTAGGA 002720 002721 CTGAGGAGCA AGCGAGCAAG CAGCAGTTCG TGGTGAAGAT AGGAAAAGAG TCCAGGAGCC AGTGCGATTT GGTGAAGGAA 002800 002801 GCTAGGAAGA AGGAAGGAGC GCTAACGATT TGGTGGTGAA GCTAGGAAAA AGGATTCCAG GAAGGAGCGA GTGCAATTTG 002880 002881 GTGATGAAGG TAGCAGGCGG CTTGGCTTGG CAACCACACG GAGGAGGCGA GCAGGCGTTG TGCGTAGAGG ATCCTAGACC 002960 002961 AGCATGCCAG TGTGCCAAGG CCACAGGGAA AGCGAGTGGT TGGTAAAAAT CCGTGAGGTC GGCAATATGT TGTTTTTCTG 003040 003041 GAACTTACTT ATGGTAACCT TTTATTTATT TTCTAATATA ATGGGGGAGT TTCGTACTGA GGTGTAAAGG GATTTATATG 003120 003121 GGGACGTAGG CCGATTTCCG GGTGTTGTAG GTTTCTCTTT TTCAGGCTTA TACTCATGAA TCTTGTCTGA AGCTTTTGAG 003200 003201 GGCAGACTGC CAAGTCCTGG AGAAATAGTA GATGGCAAGT TTGTGGGTTT TTTTTTTTTA CACGAATTTG AGGAAAACCA 003280 003281 AATGAATTTG ATAGCCAAAT TGAGACAATT TCAGCAAATC TGTAAGCAGT TTGTATGTTT AGTTGGGGTA ATGAAGTATT 003360 003361 TCAGTTTTGT GAATAGATGA CCTGTTTTTA CTTCCTCACC CTGAATTCGT TTTGTAAATG TAGAGTTTGG ATGTGTAACT 003440 003441 GAGGCGGGGG GGAGTTTTCA GTATTTTTTT TTGTGGGGGT GGGGGCAAAA TATGTTTTCA GTTCTTTTTC CCTTAGGTCT 003520 003521 GTCTAGAATC CTAAAGGCAA ATGACTCAAG GTGTAACAGA AAACAAGAAA ATCCAATATC AGGATAATCA GACCACCACA 003600 003601 GGTTTACAGT TTATAGAAAC TAGAGCAGTT CTCACGTTGA GGTCTGTGGA AGAGATGTCC ATTGGAGAAA TGGCTGGTAG 003680 003681 TTACTCTTTT TTCCCCCCAC CCCCTTAATC AGACTTTAAA AGTGCTTAAC CCCTTAAACT TGTTATTTTT TACTTGAAGC 003760 003761 ATTTTGGGAT GGTCTTAACA GGGAAGAGAG AGGGTGGGGG AGAAAATGTT TTTTTCTAAG ATTTTCCACA GATGCTATAG 003840 003841 TACTATTGAC AAACTGGGTT AGAGAAGGAG TGTACCGCTG TGCTGTTGGC ACGAACACCT TCAGGGACTG GAGCTGCTTT 003920 003921 TATCCTTGGA AGAGTATTCC CAGTTGAAGC TGAAAAGTAC AGCACAGTGC AGCTTTGGTT CATATTCAGT CATCTCAGGA 004000 004001 GAACTTCAGA AGAGCTTGAG TAGGCCAAAT GTTGAAGTTA AGTTTTCCAA TAATGTGACT TCTTAAAAGT TTTATTAAAG 004080 004081 GGGAGGGGCA AATATTGGCA ATTAGTTGGC AGTGGCCTGT TACGGTTGGG ATTGGTGGGG TGGGTTTAGG TAATTGTTTA 004160 004161 GTTTATGATT GCAGATAAAC TCATGCCAGA GAACTTAAAG TCTTAGAATG GAAAAAGTAA AGAAATATCA ACTTCCAAGT 004240 004241 TGGCAAGTAA CTCCCAATGA TTTAGTTTTT TTCCCCCCAG TTTGAATTGG GAAGCTGGGG GAAGTTAAAT ATGAGCCACT 004320 004321 GGGTGTACCA GTGCATTAAT TTGGGCAAGG AAAGTGTCAT AATTTGATAC TGTATCTGTT TTCCTTCAAA GTATAGAGCT 004400 004401 TTTGGGGAAG GAAAGTATTG AACTGGGGGT TGGTCTGGCC TACTGGGCTG ACATTAACTA CAATTATGGG AAATGCAAAA 004480 004481 GTTGTTTGGA TATGGTAGTG TGTGGTTCTC TTTTGGAATT TTTTTCAGGT GATTTAATAA TAATTTAAAA CTACTATAGA 004560 004561 AACTGCAGAG CAAAGGAAGT GGCTTAATGA TCCTGAAGGG ATTTCTTCTG ATGGTAGCTT TTGTATTATC AAGTAAGATT 004640 004641 CTATTTTCAG TTGTGTGTAA GCAAGTTTTT TTTTAGTGTA GGAGAAATAC TTTTCCATTG TTTAACTGCA AAACAAGATG 004720 004721 TTAAGGTATG CTTCAAAAAT TTTGTAAATT GTTTATTTTA AACTTATCTG TTTGTAAATT GTAACTGATT AAGAATTGTG 004800 004801 ATAGTTCAGC TTGAATGTCT CTTAGAGGGT GGGCTTTTGT TGATGAGGGA GGGGAAACTT TTTTTTTTTC TATAGACTTT 004880 004881 TTTCAGATAA CATCTTCTGA GTCATAACCA GCCTGGCAGT ATGATGGCCT AGATGCAGAG AAAACAGCTC CTTGGTGAAT 004960 004961 TGATAAGTAA AGGCAGAAAA GATTATATGT CATACCTCCA TTGGGGAATA AGCATAACCC TGAGATTCTT ACTACTGATG 005040 005041 AGAACATTAT CTGCATATGC CAAAAAATTT TAAGCAAATG AAAGCTACCA ATTTAAAGTT ACGGAATCTA CCATTTTAAA 005120 005121 GTTAATTGCT TGTCAAGCTA TAACCACAAA AATAATGAAT TGATGAGAAA TACAATGAAG AGGCAATGTC CATCTCAAAA 005200 005201 TACTGCTTTT ACAAAAGCAG AATAAAAGCG AAAAGAAATG AAAATGTTAC ACTACATTAA TCCTGGAATA AAAGAAGCCG 005280 005281 AAATAAATGA GAGATGAGTT GGGATCAAGT GGATTGAGGA GGCTGTGCTG TGTGCCAATG TTTCGTTTGC CTCAGACAGG 005360 005361 TATCTCTTCG TTATCAGAAG AGTTGCTTCA TTTCATCTGG GAGCAGAAAA CAGCAGGCAG CTGTTAACAG ATAAGTTTAA 005440 005441 CTTGCATCTG CAGTATTGCA TGTTAGGGAT AAGTGCTTAT TTTTAAGAGC TGTGGAGTTC TTAAATATCA ACCATGGCAC 005520 005521 TTTCTCCTGA CCCCTTCCCT AGGGGATTTC AGGATTGAGA AATTTTTCCA TCGAGCCTTT TTAAAATTGT AGGACTTGTT 005600 005601 CCTGTGGGCT TCAGTGATGG GATAGTACAC TTCACTCAGA GGCATTTGCA TCTTTAAATA ATTTCTTAAA AGCCTCTAAA 005680 005681 GTGATCAGTG CCTTGATGCC AACTAAGGAA ATTTGTTTAG CATTGAATCT CTGAAGGCTC TATGAAAGGA ATAGCATGAT 005760 005761 GTGCTGTTAG AATCAGATGT TACTGCTAAA ATTTACATGT TGTGATGTAA ATTGTGTAGA AAACCATTAA ATCATTCAAA 005840 005841 ATAATAAACT ATTTTTATTA GAGAATGTAT ACTTTTAGAA AGCTGTCTCC TTATTTAAAT AAAATAGTGT TTGTCTGTAG 005920 005921 TTCAGTGTTG GGGCAATCTT GGGGGGGATT CTTCTCTAAT CTTTCAGAAA CTTTGTCTGC GAACACTCTT TAATGGACCA 006000 006001 GATCAGGATT TGAGCGGAAG AACGAATGTA ACTTTAAGGC AGGAAAGACA AATTTTATTC TTCATAAAGT GATGAGCATA 006080 006081 TAATAATTCC AGGCACATGG CAATAGAGGC CCTCTAAATA AGGAATAAAT AACCTCTTAG ACAGGTGGGA GATTATGATC 006160 006161 AGAGTAAAAG GTAATTACAC ATTTTATTTC CAGAAAGTCA GGGGTCTATA AATTGACAGT GATTAGAGTA ATACTTTTTC 006240 006241 ACATTTCCAA AGTTTGCATG TTAACTTTAA ATGCTTACAA TCTTAGAGTG GTAGGCAATG TTTTACACTA TTGACCTTAT 006320 006321 ATAGGGAAGG GAGGGGGTGC CTGTGGGGTT TTAAAGAATT TTCCTTTGCA GAGGCATTTC ATCCTTCATG AAGCCATTCA 006400 006401 GGATTTTGAA TTGCATATGA GTGCTTGGCT CTTCCTTCTG TTCTAGTGAG TGTATGAGAC CTTGCAGTGA GTTTATCAGC 006480 006481 ATACTCAAAA TTTTTTTCCT GGAATTTGGA GGGATGGGAG GAGGGGGTGG GGCTTACTTG TTGTAGCTTT TTTTTTTTTT 006560 006561 ACAGACTTCA CAGAGAATGC AGTTGTCTTG ACTTCAGGTC TGTCTGTTCT GTTGGCAAGT AAATGCAGTA CTGTTCTGAT 006640 006641 CCCGCTGCTA TTAGAATGCA TTGTGAAACG ACTGGAGTAT GATTAAAAGT TGTGTTCCCC AATGCTTGGA GTAGTGATTG 006720 006721 TTGAAGGAAA AAATCCAGCT GAGTGATAAA GGCTGAGTGT TGAGGAAATT TCTGCAGTTT TAAGCAGTCG TATTTGTGAT 006800 006801 TGAAGCTGAG TACATTTTGC TGGTGTATTT TTAGGTAAAA TGCTTTTTGT TCATTTCTGG TGGTGGGAGG GGACTGAAGC 006880 006881 CTTTAGTCTT TTCCAGATGC AACCTTAAAA TCAGTGACAA GAAACATTCC AAACAAGCAA CAGTCTTCAA GAAATTAAAC 006960 006961 TGGCAAGTGG AAATGTTTAA ACAGTTCAGT GATCTTTAGT GCATTGTTTA TGTGTGGGTT TCTCTCTCCC CTCCCTTGGT 007040 007041 CTTAATTCTT ACATGCAGGA ACACTCAGCA GACACACGTA TGCGAAGGGC CAGAGAAGCC AGACCCAGTA AGAAAAAATA 007120 007121 GCCTATTTAC TTTAAATAAA CCAAACATTC CATTTTAAAT GTGGGGATTG GGAACCACTA GTTCTTTCAG ATGGTATTCT 007200 007201 TCAGACTATA GAAGGAGCTT CCAGTTGAAT TCACCAGTGG ACAAAATGAG GAAAACAGGT GAACAAGCTT TTTCTGTATT 007280 007281 TACATACAAA GTCAGATCAG TTATGGGACA ATAGTATTGA ATAGATTTCA GCTTTATGCT GGAGTAACTG GCATGTGAGC 007360 007361 AAACTGTGTT GGCGTGGGGG TGGAGGGGTG AGGTGGGCGC TAAGCCTTTT TTTAAGATTT TTCAGGTACC CCTCACTAAA 007440 007441 GGCACCGAAG GCTTAAAGTA GGACAACCAT GGAGCCTTCC TGTGGCAGGA GAGACAACAA AGCGCTATTA TCCTAAGGTC 007520 007521 AAGAGAAGTG TCAGCCTCAC CTGATTTTTA TTAGTAATGA GGACTTGCCT CAACTCCCTC TTTCTGGAGT GAAGCATCCG 007600 007601 AAGGAATGCT TGAAGTACCC CTGGGCTTCT CTTAACATTT AAGCAAGCTG TTTTTATAGC AGCTCTTAAT AATAAAGCCC 007680 007681 AAATCTCAAG CGGTGCTTGA AGGGGAGGGA AAGGGGGAAA GCGGGCAACC ACTTTTCCCT AGCTTTTCCA GAAGCCTGTT 007760 007761 AAAAGCAAGG TCTCCCCACA AGCAACTTCT CTGCCACATC GCCACCCCGT GCCTTTTGAT CTAGCACAGA CCCTTCACCC 007840 007841 CTCACCTCGA TGCAGCCAGT AGCTTGGATC CTTGTGGGCA TGATCCATAA TCGGTTTCAA GGTAACGATG GTGTCGAGGT 007920 007921 CTTTGGTGGG TTGAACTATG TTAGAAAAGG CCATTAATTT GCCTGCAAAT TGTTAACAGA AGGGTATTAA AACCACAGCT 008000 008001 AAGTAGCTCT ATTATAATAC TTATCCAGTG ACTAAAACCA ACTTAAACCA GTAAGTGGAG AAATAACATG TTCAAGAACT 008080 008081 GTAATGCTGG GTGGGAACAT GTAACTTGTA GACTGGAGAA GATAGGCATT TGAGTGGCTG AGAGGGCTTT TGGGTGGGAA 008160 008161 TGCAAAAATT CTCTGCTAAG ACTTTTTCAG GTGAACATAA CAGACTTGGC CAAGCTAGCA TCTTAGCGGA AGCTGATCTC 008240 008241 CAATGCTCTT CAGTAGGGTC ATGAAGGTTT TTCTTTTCCT GAGAAAACAA CACGTATTGT TTTCTCAGGT TTTGCTTTTT 008320 008321 GGCCTTTTTC TAGCTTAAAA AAAAAAAAAG CAAAAGATGC TGGTGGTTGG CACTCCTGGT TTCCAGGACG GGGTTCAAAT 008400 008401 CCCTGCGGCG TCTTTGCTTT GACTACTAAT CTGTCTTCAG GACTCTTTCT GTATTTCTCC TTTTCTCTGC AGGTGCTAGT 008480 008481 TCTTGGAGTT TTGGGGAGGT GGGAGGTAAC AGCACAATAT CTTTGAACTA TATACATCCT TGATGTATAA TTTGTCAGGA 008560 008561 GCTTGACTTG ATTGTATATT CATATTTACA CGAGAACCTA ATATAACTGC CTTGTCTTTT TCAGGTAATA GCCTGCAGCT 008640 008641 GGTGTTTTGA GAAGCCCTAC TGCTGAAAAC TTAACAATTT TGTGTAATAA AAATGGAGAA GCTCTAAA |
Annotation (From lncRNAdb)
| Section | Description |
|---|---|
| ID | Malat1(NR_002819.2) |
| Characteristics | Intergenic ~7kb single exon transcript (Ji (2003), Hutchinson (2007)). A conserved tRNA-like sequence at the 3' end is cleaved off and processed to generated a short tRNA-like ncRNA mascRNA (MALAT1-associated small cytoplasmic RNA) (Wilusz (2008)). Post transcriptional processing of Malat1 therefore allows two ncRNAs to be created from the one original transcript (Wilusz (2008)). |
| Expression | Both Malat1 and the processed mascRNA transcript are expressed in a wide range of tissues (Ji (2003), Hutchinson (2007), Wilusz (2008)), Bernard (2010), (Ulitsky (2011)). Malat1 is highly expressed in the brain. RNA fluorescence in-situ hybridization (RNA-FISH) on adult mouse brain sections showed high expression of nuclear-localized Malat1 transcripts in pyramidal neurons of the hippocampus, Purkinje cells of the cerebellum and neurons of the substantia nigra and motoneurons, but very low levels in non-neuronal cells (Bernard (2010)). In the hippocampus and in Purkinje cells, Malat1 was first detected between post-natal day 0 (P0) and P7 and its level increased until P28 (Bernard (2010)). This high expression appears to come from up-regulation of Malat1 during differentiation, with in-vitro differentiation of neural stem cells showing significant up-regulation of expression in neuronal and glial differentiated progeny (Mercer (2010)). Expressed in the nucleus accumbens of normal human brains and upregulated in this brain region in heroin abusers (Michelhaugh (2010)). In neuroblastoma cells Malat1 was up-regulated by the hormone oxytocin (Koshimizu (2010)). Malat1 is up-regulated in a range of cancers (Ji (2003), Yamada (2006), Lin (2007), Guffanti (2009)). High expression of Malat1 is associated with metastasis in certain histological subtypes of non-small cell lung cancer (NSCLC) and is predictive of poor prognosis (Ji (2003)). Malat1 is up-regulated in placenta previa increta/percreta, a disease characterised by excessive invasion of fetal placental trophoblasts into the uterus (Tseng (2009)). Malat1 is stable in human B cells and Hela and wt MEFs (half-life >7 hrs) but has significantly lower stability in mouse 3T3 and N2A cells. Suggesting differences in Malat1 stability both within and between species (Friedel (2009), Bernard (2010), Clark (2012), Tani (2012)). Malat1 localises to SC35 domain nuclear speckles in several cell lines (Hutchinson (2007)) (Clemson (2009)). Localisation to nuclear speckles is transcription-dependent, as RNA pol II inhibition promoted re-distribution of Malat1 ncRNA from nuclear speckles to a homogenous nuclear localisation (Bernard (2010)). Malat1 co-localises with pre-mRNA-splicing factor SF2/ASF and CC3 antigen in the nuclear speckles (Bernard (2010)). MALAT1 is enriched in nuclear speckles in interphase cells and concentrates in mitotic interchromatin granule clusters (IGCs, structural analogs of nuclear speckles present in mitotic cells), but unlike Neat1 it is not required for the structural integrity of the nuclear domain (Tripathi (2010)). mascRNA derived from Malat1 is transported to the cytoplasm (Wilusz (2008)). |
| Function | Summary: Malat1 has been found to regulate alternative splicing of endogenous target genes, and it is implicated in cancer and a series of molecular and cellular phenotypes, as indicated below. Identified as an oncogene that promotes tumorigensis. Expression of a Malat1 fragment in NIH 3T3 cells transformed cells (Li (2009)). Knockdown inhibited cell mobility and lead to decreased expression of several genes (CTHRC1, CCT4, HMMR, or ROD1) that promoted cell migration (Tano (2010)). This result may also explain why knockdown inhibited trophoblast-like cell invasion in-vitro, with MALAT1 hypothesised to regulate the level of trophoblast invasion into the uterus in vivo (Tseng (2009)). MALAT1 depletion resulted in aberrant mitosis, with a large fraction of cells accumulating at G2/M boundary, and increased cell death (Tripathi (2010)). Depletion of Malat1 in neuroblastoma cells indicated that Malat1 affects the expression of genes involved not only in the organization and the function of the nucleus, but also in synapse function and dendrite development. In cultured hippocampal neurons, knock-down of Malat1 decreased synaptic density, whereas its over-expression resulted in a cell-autonomous increase in synaptogenesis (Bernard (2010)). There is a significant enrichment for SRSF1 (SF2/ASF) binding sites within the 5' half of both human and mouse MALAT1, whose direct interaction has been demonstrated, and is dependent on its canonical RRM (RRM1) or the pseudo RRM (RRM2) domains (Sanford (2009), Tripathi (2010)). Independent sequence elements in MALAT1 influence its distribution to nuclear speckles and the recruitment of SRSF1 (Tripathi (2010)). Additional interactions have been found between MALAT1 and SRSF1, SRSF2, and SRSF3 proteins, but only weak interactions with SRSF5 and PSP1 (an RNA-binding protein that is a component of paraspeckles) have been identified (Tripathi (2010)). Knock-down of nuclear Malat1 in a transfected U2OS cell line showed that it modulates the recruitment of SR proteins (SRSF1 or SF2/ASF and SC35) to a transcriptionally active transgene array (stably integrated), indicating a role in the regulation of the association of pre-mRNA-splicing factors to transcription sites and control of post-transcriptional gene expression (Bernard (2010)). Malat1 modulates the speckle association of a subset of pre-mRNA splicing factors, such as SF1, U2AF-65, SF3a60, and B-U2snRNP (Tripathi (2010)). Malat1-depleted HeLa cells show increased cellular levels of dephosphorylated SRSF1 (SF2/ASF) (Bernard (2010)), as well as a moderately increased cytoplasmic pool of poly(A)+ RNA (Tripathi (2010)). It has recently been shown to regulate alternative splicing of endogenous target genes by modulating SR splicing factor phosphorylation, affecting their levels and the distribution and ratio of phosphorylated to dephosphorylated pools (Tripathi (2010)). Suggested to interact with the PRC2 complex in the HCT-116 cancer cell line (Guil (2012)). |
| Conservation | Therian mammals (found in oppossum as well as placental mammals) (Hutchinson (2007)). Recently described in zebrafish. Sequence homology to zebrafish was limited to the 3°Ø end, but the ~7kb long, single exon structure as well as positional synteny was conserved, as was the high expression in brain found identified in mammals (Ulitsky (2011)). |
| Name | Malat1: Metastasis-associated lung adenocarcinoma transcript 1. Neat2: Nuclear enriched abundant transcript 2 |