Difference between revisions of "NONHSAT123382"
Chunlei Yu (talk | contribs) (→Function) |
Chunlei Yu (talk | contribs) |
||
| Line 19: | Line 19: | ||
[[File:Human PINT is a p53-regulated long intergenic non-coding RNA (lincRNA) downregulated in colorectal cancer.jpg|right|thumb|400px|'''Human PINT is a p53-regulated long intergenic non-coding RNA (lincRNA) downregulated in colorectal cancer. '''<ref name="ref1" />]] | [[File:Human PINT is a p53-regulated long intergenic non-coding RNA (lincRNA) downregulated in colorectal cancer.jpg|right|thumb|400px|'''Human PINT is a p53-regulated long intergenic non-coding RNA (lincRNA) downregulated in colorectal cancer. '''<ref name="ref1" />]] | ||
| + | |||
| + | |||
In the human ortholog, PINT, is also transcriptionally regulated by p53, and its expression correlates with similar cellular pathways to those of the mouse lincRNA. However, in contrast to the mouse Pint, human PINT is a negative regulator of proliferation and survival, and is downregulated in colon cancer, representing a novel tumor suppressor candidate lincRNA<ref name="ref1" />. | In the human ortholog, PINT, is also transcriptionally regulated by p53, and its expression correlates with similar cellular pathways to those of the mouse lincRNA. However, in contrast to the mouse Pint, human PINT is a negative regulator of proliferation and survival, and is downregulated in colon cancer, representing a novel tumor suppressor candidate lincRNA<ref name="ref1" />. | ||
| Line 26: | Line 28: | ||
===Diseases=== | ===Diseases=== | ||
* colon cancer<ref name="ref1" /> | * colon cancer<ref name="ref1" /> | ||
| + | |||
| + | |||
| + | |||
===Expression=== | ===Expression=== | ||
| Line 95: | Line 100: | ||
| | AATTCAAAAACAACAGCCACAACGTCT<ref name="ref1" /> | | | AATTCAAAAACAACAGCCACAACGTCT<ref name="ref1" /> | ||
|} | |} | ||
| + | |||
| + | |||
| + | |||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
Center for Applied Medical Research, University of Navarra, 55 Pio XII Avenue., 31008 Pamplona, Spain<ref name="ref1" /> | Center for Applied Medical Research, University of Navarra, 55 Pio XII Avenue., 31008 Pamplona, Spain<ref name="ref1" /> | ||
| + | |||
| + | |||
==References== | ==References== | ||
| Line 103: | Line 113: | ||
<ref name="ref1">Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2[J]. Genome biology. 2013,14(9):R104.</ref>(1) | <ref name="ref1">Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2[J]. Genome biology. 2013,14(9):R104.</ref>(1) | ||
</references> | </references> | ||
| + | |||
| + | |||
| + | |||
{{basic| | {{basic| | ||
Revision as of 10:57, 10 April 2015
LINC-PINT, p53 induced long intergenic non-protein coding RNA, functions by regulating targeting of PRC2 to specific genes for repression
Contents
Annotated Information
Transcriptomic Nomeclature
LINC-PINT: long intergenic non-protein coding RNA, p53 induced transcript(HGNC nomenclature)
FLJ43663, LincRNA-Pint, "p53 induced noncoding transcript", PINT[1]
characteristics
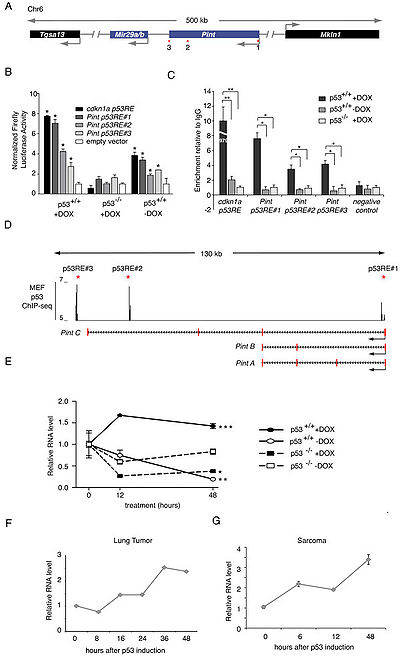
Pint is aubiquitously expressed lincRNA that is finely regulated by p53[1].
Cellular Localization
In mouse cells, Pint is a nuclear lincRNA[1].
Function
PINT is a lincRNA specifically regulated by p53 in mouse and human cells. In mouse cells, PINT promotes proliferation and survival, and functions by regulating targeting of PRC2 to specific genes for repression[1].
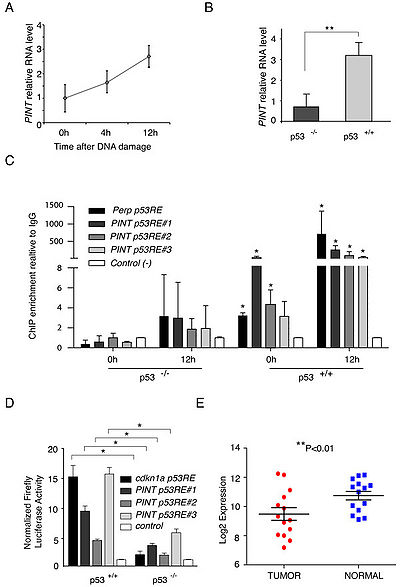
In the human ortholog, PINT, is also transcriptionally regulated by p53, and its expression correlates with similar cellular pathways to those of the mouse lincRNA. However, in contrast to the mouse Pint, human PINT is a negative regulator of proliferation and survival, and is downregulated in colon cancer, representing a novel tumor suppressor candidate lincRNA[1].
Regulation
p53[1]
Diseases
- colon cancer[1]
Expression
PINT appears to be significantly downregulated in primary colon tumors, and its overexpression in human tumor cells inhibits their proliferation.
Oligonucleotides
| Antisense Oligos(ASOs) | Sequence | Position |
|---|---|---|
| ASO#1 | GCCGACTCCCCATCTCTGCC | ex4[1] |
| ASO#2 | GGCACCCATCTCCCTTGCAA | intronic[1] |
| ASO#3 | TAGCTCAGTTCTTCTAACGC | ex3[1] |
| ASO#4 | CCTTTAGTTAGCTTTGGTCT | intronic[1] |
| ASO CTRL#1 | CCTTCCCTGAAGGTTCCTCC[1] | |
| ASO CTRL#2 | TAGTGCGGACCTACCCACGA[1] |
| siRNA | Sequence | Supplier |
|---|---|---|
| siRNA P53#1 | AGAAGAAAAUUUCCGCAAA | Ambion[1] |
| siRNA P53#2 | ACAGCGUGGUGGUACCUUA | Ambion[1] |
| siRNA control | Not available from supplier | Ambion[1] |
| shRNA cloning | Forward | Reverse |
|---|---|---|
| Ezh2 | CCGGCGGCTCCTCTAACCATGTTTACT | AATTCAAAAACGGCTCCTCTAACCATG[1] |
| Scramble | CCGGCAACAGCCACAACGTCTATATCT | AATTCAAAAACAACAGCCACAACGTCT[1] |
Labs working on this lncRNA
Center for Applied Medical Research, University of Navarra, 55 Pio XII Avenue., 31008 Pamplona, Spain[1]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2[J]. Genome biology. 2013,14(9):R104.
Basic Information
| Transcript ID |
NONHSAT123382 |
| Source |
NONCODE4.0 |
| Same with |
LINC-PINT,FLJ43663, LincRNA-Pint, PINT |
| Classification |
intergenic |
| Length |
2946 nt |
| Genomic location |
chr7-:130628919..130793562 |
| Exon number |
4 |
| Exons |
130628919..130630594,130668812..130668868,130737184..130737314,130792479..130793562 |
| Genome context |
|
| Sequence |
000001 GTCATATCTC CGTACCTCAC TTCCTGACAC AAACAAGTTT TCACTGTTGT CAGCAACAAA GCCCTAATAT AGCTGCGGAA 000080 000081 GAGAAAAACT GCATTGCATT TTGCCTCCTG CAAGCATCAT CAACAGTTAC TGGAGGAACG TAATTCCAGA AAGCTTGAAA 000160 000161 GCCGTGGTGA TGGTAATTAT GTATCAAATG CCTGGTTCTA TTTCTGTTAT TATTGTTTTG TCATTTCTGT TTTCCCAGCG 000240 000241 ATCTGACTGA ACTCGCAGAG GGACAAATCC AGTTTTTCTT TTTGACTTTT GTCAAACTAA ATCAGGCCTG ATAGAAAACT 000320 000321 CATTGCTCTC CGGGGAAACA AAGTAGGAGC CACGAAATGT CATTTTAACA GAGCGTGGGT TTGGTGACTG TAGGAAAGGA 000400 000401 TTTGAGGACG CTCCTTCTGT TCGGCTTCCT ATGTCATGAG CACAGGCTCC ACGCACGCAC AGACACCACG GCTCCCGGAT 000480 000481 GCTGTGGCTC CCCGATCGGG GCTCCTGCAG CGCCAGAAGC CCCTCCGGGA TGCTTCGAGG GGCTCCCGGT GGGTGGAGGT 000560 000561 ACGGACGCCG CTGCGGCCGC CGCCGCCAGT CCTGCTGCTG TTGTTGCTGC TGCAGTCACG TGGGAGCCCC TTTAAGTTTC 000640 000641 CATAGAGAGG CCTCTCTGGT GTCACATGAT GGACATGATA TAATGAAACA ACATTGTGGA GAGGAAAGCA TTAGGGGAGC 000720 000721 CCACGGCTAC AAAAACAAGT GAGTGAGAAG AGGTGGGAGG AAGAGAAACT ACGCCACCTC CCCTGCAGCC GAGTGCACGC 000800 000801 AGCAGCCTGG CGTGACAAGT GGGCGACGCC GGGGGGCAGG GAGCCGGGGT CCTTGGCCCT GGCCGGGGAC CCCACCGCCC 000880 000881 ACCGCGCGGA GGACAACTTT TAGCCGGCAG CCCAGACCAG CGCGGCACCT GTCTCCGGAG TCTCCACCGC TCCTCCCGAT 000960 000961 TCATCCCAGG GAAATTCTCA AGAATACGCT CTACAAATCT ACGTGCGCAT CATTTTCACC TCGCGTCGCG CCCGGGAGGA 001040 001041 AGGAACGAGG CAAGGAGCTA AAGCAGCGTG CGTTCAGCCC TGGGGCATTT TATTAATGCT TTTACGAGTT AGAAGAGTTG 001120 001121 GGATAATTTG CCATCTGGAG TTTCTCTGCC TTGCTGATCT GAGCTCAGAC CTGCCAATTT ACCAGAGATA ATTGATAACA 001200 001201 CCCTGTAACA GCTGAGAGGA AAATGGAAGA AACGGAGATA CTTTTAGTGA AGCAGAATAA ACCACTGAAC AGGAAAAATG 001280 001281 AGGAAGCTGT GAGTACCAGT GGAAGGAACG AGCCAGGAAG AGGGACTGGA ACCATCTCAG AAGCCATGCC CCTCAGGCTG 001360 001361 GAACTTGCCC TGTCTCCTCG CAGATGAGGT AGGAGGCTCA GCACGGGCTG GTGGGAGCAG CCCACAGCAG CAGTGAGGGT 001440 001441 CAGTGGGCCT GTGAGTTAAG AAGAAGGTGG CACAGAGCGA GGGTCTCTGG ATCCTGACTG TTTGACTTTT CCATGATTAG 001520 001521 GATAAGTAGC CAGGGCTTGG CTAGTTGGAG AGTTACTCGA ACCTCAGGTG ACAGTTGTAA GGCAGCACAT AGTGAAAAAG 001600 001601 AGTCCTAGCC TGGGAAAGTC CAAAACCTTA GGTCTGGTTT CAGTTCACTC ACCTATCTCT GTGAC |