Difference between revisions of "NONHSAT017465"
(→Disease) |
(→Disease) |
||
| Line 72: | Line 72: | ||
thyroid cancer<ref name="ref8" /> | thyroid cancer<ref name="ref8" /> | ||
| + | |||
You can also add sub-section(s) at will. | You can also add sub-section(s) at will. | ||
Revision as of 11:54, 30 June 2017
Contents
Annotated Information
Name
H19: imprinted maternally expressed transcript (non-protein coding)
ASM, ASM1, D11S813E, LINC00008, "long intergenic non-protein coding RNA 8", NCRNA00008, "non-protein coding RNA 8"
Cellular Localization
H19 RNA is cytoplasmic but not associated with the translational machinery.[1]
Instead, it is located in a particle with a sedimentation coefficient of approximately 28S. [1]
Conservation
The H19 gene is conserved in chicken. [1]
Conserved and imprinted in therians (eutherians and marsupials). Both the miRNA and the H19 lncRNA exon structure are conserved.[2]
Function
H19 knockdown activates SAHH, leading to increased DNMT3B-mediated methylation of an lncRNA-encoding gene Nctc1 within the Igf2-H19-Nctc1 locus.[3]
H19 acts as an oncogene and promotes ESCC cell proliferation and metastasis, which may infer H19 as a potential therapeutic target for treating ESC patients.[4]
H19 contributes to cardiac fibroblast proliferation and fibrosis, which act in part through repression of DUSP5/ERK1/2.[5]
The overexpression of H19 might potentially serve as a reliable biomarker for poor prognosis in different types of cancers.[6]
H19 long noncoding RNA alters trophoblast cell migration and invasion by regulating TβR3 in placentae with fetal growth restriction.[7]
H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer, whose mechanism contributes to a better understanding of thyroid cancer pathogenesis. [8]
H19 interference in GBC suppressed tumor cell invasion and promoted mesenchymal-epithelial transition (MET) via suppressing Twist expression.[9]
H19 potentiated Wnt/β-catenin pathway by serving as a molecular sponge for miR-141 and miR-22, leading to the promotion of osteoblast differentiation. [10]
Expression
| Primer | Forward primer | Reverse primer |
|---|---|---|
| RT-PCR | 5'-CCTCAAGATGAAAGAAATGGTGCTA-3' | 5'-TCCATCTCCCTTGCTGTATC -3' [10] |
| ChIP PCR primer sequences | 5'-CCAAAAGTAACCGGGATGAA-3' | 5'-CTGGAGTCTGGCAGGAATGT -3' [10] |
| T7 PCR primer sequences | 5'-TAATACGACTCACTATAGGGCTTCTTTTTCATCCTT-3' | 5'-CAAATGACTTAGTGCAAATTAAATTCAGAAGGGAC-3'[10] |
Regulation
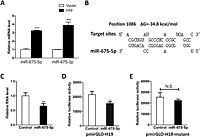
H19 is negatively regulated by miR-675-5p. [10]
Allelic Information and Variation
Please input allelic information and variation information here.
Evolution
Please input evolution information here.
Disease
gallbladder carcinoma[9]
esophageal squamous cell carcinoma[4]
thyroid cancer[8]
You can also add sub-section(s) at will.
Labs working on this lncRNA
- Howard Hughes Medical Institute, Princeton University New Jersey 08544.[1]
- The Babraham Institute, Laboratory of Developmental Genetics and Imprinting, Cambridge CB22 3AT, UK.[2]
- Department of Obstetrics, Gynecology, and Reproductive Sciences, Yale School of Medicine, New Haven, Connecticut 06510, USA. [3]
- Department of Cardiothoracic Surgery, Southwest Hospital. [4]
- Department of Cardiothoracic Surgery, The Second Hospital of Anhui Medical University, Hefei 230601, China.[5]
- The Second Clinical Medical College of Nanjing Medical University, Nanjing, China[6]
- Department of Obstetrics, Gynecology and Reproductive Sciences, Yale University School of Medicine, New Haven, CT, USA[7]
- Department of Nuclear Medicine, Shanghai Tenth people's hospital, Tongji university, Shanghai, China. [8]
- Department of General Surgery, Xinhua Hospital, Shanghai Jiao Tong University School of Medicine Shanghai 200000, China. [9]
- School of Biomedical Sciences, The Chinese University of Hong Kong, Shatin, New Territories, Hong Kong, P.R. China. [10]
References
- ↑ 1.0 1.1 1.2 1.3 Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA[J]. Molecular and cellular biology. 1990,10(1):28-36.
- ↑ 2.0 2.1 Smits G, Mungall AJ, Griffiths-Jones S, Smith P, Beury D, Matthews L, et al. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians[J]. Nature genetics. 2008,40(8):971-6.
- ↑ 3.0 3.1 Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N, et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase[J]. Nature communications. 2015,6:10221.
- ↑ 4.0 4.1 4.2 Tan D, Wu Y, Hu L, He P, Xiong G, Bai Y, et al. Long noncoding RNA H19 is up-regulated in esophageal squamous cell carcinoma and promotes cell proliferation and metastasis[J]. Diseases of the esophagus : official journal of the International Society for Diseases of the Esophagus / ISDE. 2016.
- ↑ 5.0 5.1 Tao H, Cao W, Yang JJ, Shi KH, Zhou X, Liu LP, et al. Long noncoding RNA H19 controls DUSP5/ERK1/2 axis in cardiac fibroblast proliferation and fibrosis[J]. Cardiovascular pathology : the official journal of the Society for Cardiovascular Pathology. 2016,25(5):381-9.
- ↑ 6.0 6.1 Chen T, Yang P, He ZY. Long non-coding RNA H19 can predict a poor prognosis and lymph node metastasis: a meta-analysis in human cancer[J]. Minerva medica. 2016,107(4):251-8.
- ↑ 7.0 7.1 Zuckerwise L, Li J, Lu L, Men Y, Geng T, Buhimschi CS, et al. H19 long noncoding RNA alters trophoblast cell migration and invasion by regulating TbetaR3 in placentae with fetal growth restriction[J]. Oncotarget. 2016.
- ↑ 8.0 8.1 8.2 Liu L, Yang J, Zhu X, Li D, Lv Z, Zhang X. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer[J]. The FEBS journal. 2016,283(12):2326-39.
- ↑ 9.0 9.1 9.2 Wang SH, Wu XC, Zhang MD, Weng MZ, Zhou D, Quan ZW. Upregulation of H19 indicates a poor prognosis in gallbladder carcinoma and promotes epithelial-mesenchymal transition[J]. American journal of cancer research. 2016,6(1):15-26.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 Liang WC, Fu WM, Wang YB, Sun YX, Xu LL, Wong CW, et al. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA[J]. Scientific reports. 2016,6:20121.
Basic Information
| Transcript ID |
NONHSAT017465 |
| Source |
NONCODE4.0 |
| Same with |
, |
| Classification |
intergenic |
| Length |
2322 nt |
| Genomic location |
chr11-:2016406..2019105 |
| Exon number |
5 |
| Exons |
2016406..2017024,2017106..2017228,2017309..2017421,2017517..2017651,2017748..2019105 |
| Genome context |
|
| Sequence |
000001 GGGAGGGGGT GGGATGGGTG GGGGGTAACG GGGGAAACTG GGGAAGTGGG GAACCGAGGG GCAACCAGGG GAAGATGGGG 000080
000081 TGCTGGAGGA GAGCTTGTGG GAGCCAAGGA GCACCTTGGA CATCTGGAGT CTGGCAGGAG TGATGACGGG TGGAGGGGCT 000160 000161 AGCTCGAGGC AGGGCTGGTG GGGCCTGAGG CCAGTGAGGA GTGTGGAGTA GGCGCCCAGG CATCGTGCAG ACAGGGCGAC 000240 000241 ATCAGCTGGG GACGATGGGC CTGAGCTAGG GCTGGAAAGA AGGGGGAGCC AGGCATTCAT CCCGGTCACT TTTGGTTACA 000320 000321 GGACGTGGCA GCTGGTTGGA CGAGGGGAGC TGGTGGGCAG GGTTTGATCC CAGGGCCTGG GCAACGGAGG TGTAGCTGGC 000400 000401 AGCAGCGGGC AGGTGAGGAC CCCATCTGCC GGGCAGGTGA GTCCCTTCCC TCCCCAGGCC TCGCTTCCCC AGCCTTCTGA 000480 000481 AAGAAGGAGG TTTAGGGGAT CGAGGGCTGG CGGGGAGAAG CAGACACCCT CCCAGCAGAG GGGCAGGATG GGGGCAGGAG 000560 000561 AGTTAGCAAA GGTGACATCT TCTCGGGGGG AGCCGAGACT GCGCAAGGCT GGGGGGTTAT GGGCCCGTTC CAGGCAGAAA 000640 000641 GAGCAAGAGG GCAGGGAGGG AGCACAGGGG TGGCCAGCGT AGGGTCCAGC ACGTGGGGTG GTACCCCAGG CCTGGGTCAG 000720 000721 ACAGGGACAT GGCAGGGGAC ACAGGACAGA GGGGTCCCCA GCTGCCACCT CACCCACCGC AATTCATTTA GTAGCAGGCA 000800 000801 CAGGGGCAGC TCCGGCACGG CTTTCTCAGG CCTATGCCGG AGCCTCGAGG GCTGGAGAGC GGGAAGACAG GCAGTGCTCG 000880 000881 GGGAGTTGCA GCAGGACGTC ACCAGGAGGG CGAAGCGGCC ACGGGAGGGG GGCCCCGGGA CATTGCGCAG CAAGGAGGCT 000960 000961 GCAGGGGCTC GGCCTGCGGG CGCCGGTCCC ACGAGGCACT GCGGCCCAGG GTCTGGTGCG GAGAGGGCCC ACAGTGGACT 001040 001041 TGGTGACGCT GTATGCCCTC ACCGCTCAGC CCCTGGGGCT GGCTTGGCAG ACAGTACAGC ATCCAGGGGA GTCAAGGGCA 001120 001121 TGGGGCGAGA CCAGACTAGG CGAGGCGGGC GGGGCGGAGT GAATGAGCTC TCAGGAGGGA GGATGGTGCA GGCAGGGGTG 001200 001201 AGGAGCGCAG CGGGCGGCGA GCGGGAGGCA CTGGCCTCCA GAGCCCGTGG CCAAGGCGGG CCTCGCGGGC GGCGACGGAG 001280 001281 CCGGGATCGG TGCCTCAGCG TTCGGGCTGG AGACGAGGCC AGGTCTCCAG CTGGGGTGGA CGTGCCCACC AGCTGCCGAA 001360 001361 GGCCAAGACG CCAGGTCCGG TGGACGTGAC AAGCAGGACA TGACATGGTC CGGTGTGACG GCGAGGACAG AGGAGGCGCG 001440 001441 TCCGGCCTTC CTGAACACCT TAGGCTGGTG GGGCTGCGGC AAGAAGCGGG TCTGTTTCTT TACTTCCTCC ACGGAGTCGG 001520 001521 CACACTATGG CTGCCCTCTG GGCTCCCAGA ACCCACAACA TGAAAGAAAT GGTGCTACCC AGCTCAAGCC TGGGCCTTTG 001600 001601 AATCCGGACA CAAAACCCTC TAGCTTGGAA ATGAATATGC TGCACTTTAC AACCACTGCA CTACCTGACT CAGGAATCGG 001680 001681 CTCTGGAAGG TGAAGCTAGA GGAACCAGAC CTCATCAGCC CAACATCAAA GACACCATCG GAACAGCAGC GCCCGCAGCA 001760 001761 CCCACCCCGC ACCGGCGACT CCATCTTCAT GGCCACCCCC TGCGGCGGAC GGTTGACCAC CAGCCACCAC ATCATCCCAG 001840 001841 AGCTGAGCTC CTCCAGCGGG ATGACGCCGT CCCCACCACC TCCCTCTTCT TCTTTTTCAT CCTTCTGTCT CTTTGTTTCT 001920 001921 GAGCTTTCCT GTCTTTCCTT TTTTCTGAGA GATTCAAAGC CTCCACGACT CTGTTTCCCC CGTCCCTTCT GAATTTAATT 002000 002001 TGCACTAAGT CATTTGCACT GGTTGGAGTT GTGGAGACGG CCTTGAGTCT CAGTACGAGT GTGCGTGAGT GTGAGCCACC 002080 002081 TTGGCAAGTG CCTGTGCAGG GCCCGGCCGC CCTCCATCTG GGCCGGGTGA CTGGGCGCCG GCTGTGTGCC CGAGGCCTCA 002160 002161 CCCTGCCCTC GCCTAGTCTG GAAGCTCCGA CCGACATCAC GGAGCAGCCT TCAAGCATTC CATTACGCCC CATCTCGCTC 002240 002241 TGTGCCCCTC CCCACCAGGG CTTCAGCAGG AGCCCTGGAC TCATCATCAA TAAACACTGT TACAGCAAAA AAAAAAAAAA 002320 002321 AA |