Difference between revisions of "PLUT"
(→Expression) |
(→Function) |
||
| Line 12: | Line 12: | ||
''PLUTO'' Regulates ''PDX1'', an essential transcriptional regulator of pancreas development and β cell function that has been implicated in genetic mechanisms underlying Mendelian and type 2 diabetes <ref name="ref1" />. | ''PLUTO'' Regulates ''PDX1'', an essential transcriptional regulator of pancreas development and β cell function that has been implicated in genetic mechanisms underlying Mendelian and type 2 diabetes <ref name="ref1" />. | ||
| − | Both PLUTO and PDX1 are downregulated in islets from donors with type 2 diabetes or impaired glucose tolerance <ref name="ref1" />. | + | Both ''PLUTO'' and ''PDX1'' are downregulated in islets from donors with type 2 diabetes or impaired glucose tolerance <ref name="ref1" />. |
===Expression=== | ===Expression=== | ||
Latest revision as of 04:30, 26 November 2018
PLUT is a long noncoding RNA located upstream of pancreatic and duodenal homeobox 1 (PDX1) and is invovled in Type 2 diabetes.
Contents
Annotated Information
Name
PLUT: PDX1 associated lncRNA, upregulator of transcription (HGNC nomenclature), PLUTO [1]
Characteristics
PLUT is a multi-isoform transcript that contains five major exons and span nearly 100 kb on human chromosome 13q12.2 (HGNC), encompassing a cluster of enhancers that make 3D contacts with the PDX1 promoter in human islets and in EndoC-βH1 cells [1].
Function
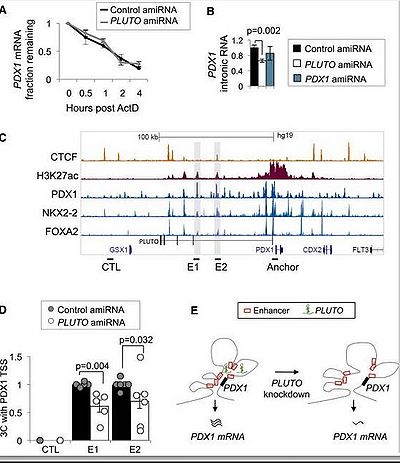
PLUTO is involved in the regulation of β cell-specific transcription factor networks [1]. It is found that PLUTO affects local 3D chromatin structure and transcription of PDX1 (encoding a key β cell transcription factor), and knockdown of PLUTO RNA in dispersed primary human islet cells causes decreased PDX1 mRNA [1].
PLUTO Regulates PDX1, an essential transcriptional regulator of pancreas development and β cell function that has been implicated in genetic mechanisms underlying Mendelian and type 2 diabetes [1]. Both PLUTO and PDX1 are downregulated in islets from donors with type 2 diabetes or impaired glucose tolerance [1].
Expression
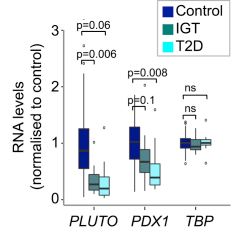
PLUTO is the most markedly downregulated lncRNAs in islets from T2D (type 2 diabetes) or IGT (impaired glucose tolerance) donors [1].
Diseases
Labs working on this lncRNA
- Section of Epigenomics and Disease, Department of Medicine, Imperial College London, London W12 0NN, United Kingdom.[1]
- Genomic Programming of Beta Cells Laboratory, Institut d'Investigacions Biomediques August Pi I Sunyer (IDIBAPS), Barcelona 08036, Spain.[1]
- Centro de Investigación Biomédica en Red de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM), Madrid 28029, Spain.[1]
- Department of Genetics and Genomic Science, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.[1]
- Sorbonne Universités, UPMC Univ Paris 06, INSERM, CNRS, Institut du cerveau et de la moelle (ICM) – Hôpital Pitié-Salpêtrière, Boulevard de l’Hôpital, Paris 75013, France.[1]
- Germans Trias i Pujol University Hospital and Research Institute and Josep Carreras Leukaemia Research Institute, Badalona 08916, Spain.[1]
- Diabetes, Obesity, and Metabolism Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.[1]
- European Genomic Institute for Diabetes, INSERM UMR 1190, Lille 59800, France.[1]
- Diabetes Research Institute (HSR-DRI), San Raffaele Scientific Institute, Milano 20132, Italy.[1]
- Cell Isolation and Transplantation Center, University of Geneva, 1211 Geneva 4, Switzerland.[1]
- Department of Clinical Sciences, Lund University Diabetes Centre, Lund University, Lund 20502, Sweden.[1]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 Akerman I, Tu Z, Beucher A, Rolando DM, Sauty-Colace C, Benazra M, Nakic N, Yang J, Wang H, Pasquali L, Moran I, Garcia-Hurtado J, Castro N, Gonzalez-Franco R, Stewart AF, Bonner C, Piemonti L, Berney T, Groop L, Kerr-Conte J, Pattou F, Argmann C, Schadt E, Ravassard P, Ferrer J. Human Pancreatic β Cell lncRNAs Control Cell-Specific Regulatory Networks. Cell Metab. 2017 Feb 7;25(2):400-411.
Sequence
>gi|100861550|ref|NR_047484.2| Homo sapiens PDX1 associated lncRNA, upregulator of transcription (PLUT), long non-coding RNA
000081 CCCCCTTTCC CTTCACAGAG ACGCGGTTTA CACCCGGGAG AACACAGGTT ACCTTGTTCT TGCTGGACCT TCCACAAGTG 000160
000161 TTTGCACATG ACCTGGAAGA CAGTCAGAGG CAGAAGTCCC TGCAAATGCT CCTGGCTACT TCCTGCAGGC AGCTCATGCT 000240
000241 GCTGCAGCAG AAGACTTTCC TATGTCTAGC TGCCAGTATT CTGGGTGCTG GGGGCATCCT GTTCCTAGGA TCACAGCCAT 000320
000321 GGTGGTATGT TCTAGAACTC AGAAATTTAC CAAATCACTG GATGGACCAG AGGAGTGAGC TCCAGGTTGG GTTTCCAGAA 000400
000401 GGGACTCCCA GAACTCTTCC ACCATACAGG CCTCTCAGGG AGCTGCTCCC TCTGCCATGG TCAGTGAGAG GGGGAAGCAG 000480
000481 GAGCCGCCAT TGGGGTTGTT GAGTTCGTGG CTGCAACCCA GGGATGAGGA AGCTGCTGCT ACACACCCAT GAAGCTGATG 000560
000561 CCTGGACATA AATCCCTACT GATAAGTGTT TACGACATTT CCAGCGTGGT GCCGACACTG CATGGAAATG CTGCATGGAA 000640
000641 AGTCCTTATA CATCTATCTT TGTGCATTTA TTGTGAGCAC CTACTATGAG TAAAACCTGG GCTGGTGGCT GGAGAAACAT 000720
000721 GAAGATGAGT AAGAGCCAAT TCCTGTTCTT GGGGATTTAA TAATATATTC AAGGGAAAAG ACACAAAATA ACCATTTCCA 000800
000801 GGATTCAAGT ATTTTGTTTC GTGCGCATTT CCAACCCATC AAATCAGCTG CAGGCACCGC ACCGTCTAAC AGACCCAGGT 000880
000881 GTTCCTGCTG GAAGAAACTG TCTTTAAGTT TCCCAGAATT GAAGGGGCTT TTCTCGCTTG CTCACGCCCA CTCTTTTGAA 000960
000961 ACCCTAATGG CTGAAGAGCA TCATCAAGAG AAGGGGAAGA TGAAGCTGAC TCCCTGCACC TGCTTCCTCC CCATTCTGTT 001040
001041 GGCTCGGGTC GGTGTGGGCT TTTAGCCTCT CTGAATGTGG AGGTTCCCAG GGATACGGGG AACCACAAAG GTCCATATTC 001120
001121 TCAAATGCAG CACTTCTTAA CCAAGGCATG GCTTTTTCTA GGTGGTCGGA GGCCAAATAC TCTGCTGCTG GCCATCCCAA 001200
001201 CCAGTAGGCA CAGGGAAATG CCAGAAAAGA AGCAGCTGGG GCTTCTCTTG CTGAGGAAGA GGGTGAAATG AACCTTGCTT 001280
001281 GCCGCAGCCA GTTTCTGACC CTGCTGCAGA GATGACGGAT GAGATTCCAG CTGTGATGGA GATGTGGTCC TGGGAGGCCA 001360
001361 TTCTAGGCAC TGCTCTTCAA AGAGGTTGTC TCCATCCTGG GATAGAAGTT CATAAGGGGA AATGTACAAA CACAGTTTTC 001440
001441 CTGCCACCTC CAAGCTGCTT GGGGCCGGAC TGGATGCCCT GCCGGCCCCT CACTCTTCTT ACCTTTGAAC ATCGACCCTG 001520
001521 GCCAGGCATT TGTTTAGGCA GGTTTTCTAA TTTAACTTTT ACAACAACCT GTGAAATGGC TACGATTAAC CCGCTTTCAT 001600
001601 GGCTGAGCAC CTGGCTGGGA GATGTAAGGT ACTTGCCCAA GGAAACCAAT CAATTATCAA CCTGAATTTG CCTGACTTCC 001680
001681 CACTCATGCT TTGGTTGCAG TTCCAGAGTC TGCTACAGGA TCTCCCATAC AGAGTGGCCT GAAGGCACTG GTTTCCCTTG 001760
001761 GGAAACAGCG AACTCATGTC CCTGGACTTC TCCCCTGTGG CCTCCCTCCC TGCTTCCCTG GGACTGTTAG TACCTTCTCC 001840
001841 TATGTTTCAT GTTGACCATA CAGCTTTGCC TTTTTCCAGT CTAGGTTTAT GCCAGACATT GTGTCTGTTA TTTTCCCTTT 001920
001921 TTAAAGTGAT ATCTGGGCTG AGTG