Difference between revisions of "DDX11-AS1"
Qianpeng Li (talk | contribs) |
|||
| Line 3: | Line 3: | ||
DDX11-AS1 (DDX11 antisense RNA 1) | DDX11-AS1 (DDX11 antisense RNA 1) | ||
===Synonyms=== | ===Synonyms=== | ||
| − | CONCR | + | CONCR, SCAT4 |
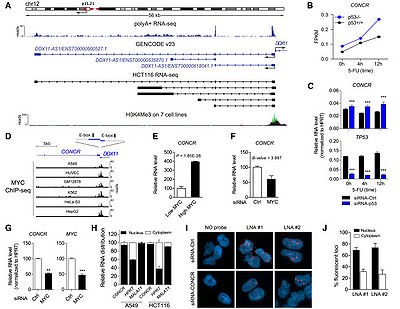
[[File:CONCR Is a Nuclear lncRNA Negatively Regulated by p53 and Activated by MYC.jpg|right|thumb|400px|'''DDX11-AS1 is a nuclear lncRNA negatively regulated by p53 and activated by MYC'''<ref name="ref1" />]] | [[File:CONCR Is a Nuclear lncRNA Negatively Regulated by p53 and Activated by MYC.jpg|right|thumb|400px|'''DDX11-AS1 is a nuclear lncRNA negatively regulated by p53 and activated by MYC'''<ref name="ref1" />]] | ||
[[File:CONCR Is Upregulated in Multiple Cancer Types.jpg|right|thumb|400px|'''DDX11-AS1 is upregulated in multiple cancer types'''<ref name="ref1" />]] | [[File:CONCR Is Upregulated in Multiple Cancer Types.jpg|right|thumb|400px|'''DDX11-AS1 is upregulated in multiple cancer types'''<ref name="ref1" />]] | ||
| Line 10: | Line 10: | ||
===RefSeq(supplied by NCBI)=== | ===RefSeq(supplied by NCBI)=== | ||
NR_038927 | NR_038927 | ||
| + | ===LncBook transcript ID=== | ||
| + | HSALNT0288928 | ||
===Characteristics=== | ===Characteristics=== | ||
DDX11-AS1 is a divergent non-overlapping transcript of the protein-coding gene DDX11, which is a predominant nuclear lncRNA. ChIP-seq data from ENCODE shows that MYC is bound to DDX11-AS1 promoter region in multiple cell types. | DDX11-AS1 is a divergent non-overlapping transcript of the protein-coding gene DDX11, which is a predominant nuclear lncRNA. ChIP-seq data from ENCODE shows that MYC is bound to DDX11-AS1 promoter region in multiple cell types. | ||
===Expression=== | ===Expression=== | ||
DDX11-AS1 is expressed in a panel of different human cell lines and upregulated in multiple cancer types. <ref name="ref1" /> | DDX11-AS1 is expressed in a panel of different human cell lines and upregulated in multiple cancer types. <ref name="ref1" /> | ||
| + | |||
| + | The temporal expression of SCAT4, SCAT5, SCAT7, and SCAT8 elevated during S-phase in serum-starved Caki-2 cells<ref name="ref2" />. | ||
| + | |||
===Regulated=== | ===Regulated=== | ||
DDX11-AS1 is negatively regulated by p53 and activated by MYC. <ref name="ref1" /> | DDX11-AS1 is negatively regulated by p53 and activated by MYC. <ref name="ref1" /> | ||
| Line 21: | Line 26: | ||
Depletion of DDX11-AS1 causes sister chromatid cohesion defects | Depletion of DDX11-AS1 causes sister chromatid cohesion defects | ||
DDX11-AS1 interacts with DDX11 and regulates its function through enhancing the ATPase activity of DDX11.<ref name="ref1" /> | DDX11-AS1 interacts with DDX11 and regulates its function through enhancing the ATPase activity of DDX11.<ref name="ref1" /> | ||
| + | |||
| + | Depletion of the selected SCATs including SCAT4 in Caki-2 cells altered cell proliferation, inhibited cell cycle progression, and induced apoptosis <ref name="ref2" />. | ||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
* Center for Applied Medical Research (CIMA), Department of Gene Therapy and Regulation of Gene Expression, University of Navarra, 55 Pio XII Avenue, 31008 Pamplona, Spain; Institute of Health Research of Navarra (IdiSNA), 31008 Pamplona, Spain. <ref name="ref1" /> | * Center for Applied Medical Research (CIMA), Department of Gene Therapy and Regulation of Gene Expression, University of Navarra, 55 Pio XII Avenue, 31008 Pamplona, Spain; Institute of Health Research of Navarra (IdiSNA), 31008 Pamplona, Spain. <ref name="ref1" /> | ||
*Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, NIH Biomedical Research Center, 251 Bayview Boulevard, Baltimore, MD 21224, USA. <ref name="ref1" /> | *Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, NIH Biomedical Research Center, 251 Bayview Boulevard, Baltimore, MD 21224, USA. <ref name="ref1" /> | ||
*Center for Applied Medical Research (CIMA), Department of Gene Therapy and Regulation of Gene Expression, University of Navarra, 55 Pio XII Avenue, 31008 Pamplona, Spain; Institute of Health Research of Navarra (IdiSNA), 31008 Pamplona, Spain. <ref name="ref1" /> | *Center for Applied Medical Research (CIMA), Department of Gene Therapy and Regulation of Gene Expression, University of Navarra, 55 Pio XII Avenue, 31008 Pamplona, Spain; Institute of Health Research of Navarra (IdiSNA), 31008 Pamplona, Spain. <ref name="ref1" /> | ||
| + | |||
==References== | ==References== | ||
<references> | <references> | ||
<ref name="ref1"> Marchese, F.P., et al., A Long Noncoding RNA Regulates Sister Chromatid Cohesion. Mol Cell, 2016. 63(3): p. 397-407. | <ref name="ref1"> Marchese, F.P., et al., A Long Noncoding RNA Regulates Sister Chromatid Cohesion. Mol Cell, 2016. 63(3): p. 397-407. | ||
</ref>(1) | </ref>(1) | ||
| + | <ref name="ref2"> | ||
| + | Ali MM, Akhade VS, Kosalai ST, Subhash S, Statello L, Meryet-Figuiere M, Abrahamsson J, Mondal T, Kanduri C. PAN-cancer analysis of S-phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nat Commun. 2018 Feb 28;9(1):883. | ||
| + | </ref>(2) | ||
</references> | </references> | ||
Revision as of 13:50, 7 August 2019
Contents
Annotated Information
Approved Symbol
DDX11-AS1 (DDX11 antisense RNA 1)
Synonyms
CONCR, SCAT4

Chromosome
12p11.21
RefSeq(supplied by NCBI)
NR_038927
LncBook transcript ID
HSALNT0288928
Characteristics
DDX11-AS1 is a divergent non-overlapping transcript of the protein-coding gene DDX11, which is a predominant nuclear lncRNA. ChIP-seq data from ENCODE shows that MYC is bound to DDX11-AS1 promoter region in multiple cell types.
Expression
DDX11-AS1 is expressed in a panel of different human cell lines and upregulated in multiple cancer types. [1]
The temporal expression of SCAT4, SCAT5, SCAT7, and SCAT8 elevated during S-phase in serum-starved Caki-2 cells[2].
Regulated
DDX11-AS1 is negatively regulated by p53 and activated by MYC. [1]
Function
DDX11-AS1 contributes to tumor growth. [1] Expression of DDX11-AS1 is periodic in the cell cycle and its presence is required for efficient G1/S transition and DNA replication.[1] Depletion of DDX11-AS1 causes sister chromatid cohesion defects DDX11-AS1 interacts with DDX11 and regulates its function through enhancing the ATPase activity of DDX11.[1]
Depletion of the selected SCATs including SCAT4 in Caki-2 cells altered cell proliferation, inhibited cell cycle progression, and induced apoptosis [2].
Labs working on this lncRNA
- Center for Applied Medical Research (CIMA), Department of Gene Therapy and Regulation of Gene Expression, University of Navarra, 55 Pio XII Avenue, 31008 Pamplona, Spain; Institute of Health Research of Navarra (IdiSNA), 31008 Pamplona, Spain. [1]
- Laboratory of Molecular Gerontology, National Institute on Aging, National Institutes of Health, NIH Biomedical Research Center, 251 Bayview Boulevard, Baltimore, MD 21224, USA. [1]
- Center for Applied Medical Research (CIMA), Department of Gene Therapy and Regulation of Gene Expression, University of Navarra, 55 Pio XII Avenue, 31008 Pamplona, Spain; Institute of Health Research of Navarra (IdiSNA), 31008 Pamplona, Spain. [1]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Marchese, F.P., et al., A Long Noncoding RNA Regulates Sister Chromatid Cohesion. Mol Cell, 2016. 63(3): p. 397-407.
- ↑ 2.0 2.1 Ali MM, Akhade VS, Kosalai ST, Subhash S, Statello L, Meryet-Figuiere M, Abrahamsson J, Mondal T, Kanduri C. PAN-cancer analysis of S-phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nat Commun. 2018 Feb 28;9(1):883.