Difference between revisions of "MIR122HG"
| Line 2: | Line 2: | ||
===Name=== | ===Name=== | ||
MIR122HG | MIR122HG | ||
| − | [[File:MIR122HG.jpg|right|thumb|400px|''' | + | [[File:MIR122HG.jpg|right|thumb|400px|'''MIR122HG transcripts are capped but not polyadenylated'''<ref name="ref1" />.]] |
===Alias=== | ===Alias=== | ||
| Line 12: | Line 12: | ||
===Characteristic=== | ===Characteristic=== | ||
| − | + | MIR122HG transcripts are capped but not polyadenylated<ref name="ref1" /> | |
| − | + | MIR122HG 3' end is generated by Drosha cleavage and not by CPA<ref name="ref1" /> | |
Pol II transcribing lnc-pri-miR-122 fails to recognize PAS (polyadenylation site) even when Microprocessor cleavage is inhibited.<ref name="ref1" /> | Pol II transcribing lnc-pri-miR-122 fails to recognize PAS (polyadenylation site) even when Microprocessor cleavage is inhibited.<ref name="ref1" /> | ||
===Expression=== | ===Expression=== | ||
| − | + | MIR122HG transcripts are capped but not polyadenylated<ref name="ref1" /> | |
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
Revision as of 06:03, 10 August 2019
Contents
Annotated Information
Name
MIR122HG
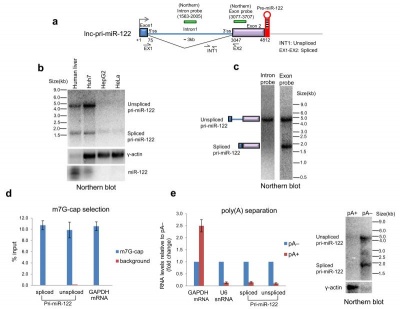
MIR122HG transcripts are capped but not polyadenylated[1].
Alias
lnc-pri-miR-122
ID
ENSG00000267391
locus
18q21.31
Characteristic
MIR122HG transcripts are capped but not polyadenylated[1] MIR122HG 3' end is generated by Drosha cleavage and not by CPA[1] Pol II transcribing lnc-pri-miR-122 fails to recognize PAS (polyadenylation site) even when Microprocessor cleavage is inhibited.[1]
Expression
MIR122HG transcripts are capped but not polyadenylated[1]
Labs working on this lncRNA
- Sir William Dunn School of Pathology, University of Oxford, Oxford, UK.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Dhir A, Dhir S, Proudfoot NJ, Jopling CL. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol. 2015 Apr;22(4):319-27. doi: 10.1038/nsmb.2982. Epub 2015 Mar 2. PubMed PMID: 25730776; PubMed Central PMCID: PMC4492989.