Difference between revisions of "Main Page"
| Line 37: | Line 37: | ||
|valign="top" style="border:1px solid #66B3FF; background-color:transparent; width:40%;"| | |valign="top" style="border:1px solid #66B3FF; background-color:transparent; width:40%;"| | ||
<div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Data sources</div> | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Data sources</div> | ||
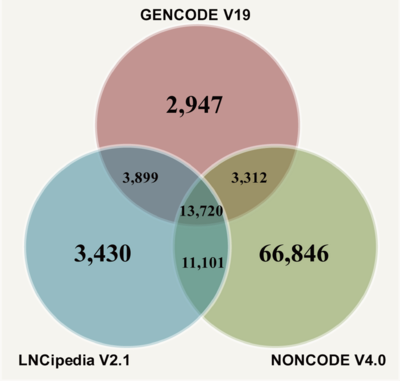
| − | <div style="padding:0.5em 0.5em;">LncRNAs in present database are collected from GENCODE | + | <div style="padding:0.5em 0.5em;">LncRNAs in present database are collected from GENCODE V19, NONCODE V4, and LNCipedia V2.1. |
| − | |||
<br /> | <br /> | ||
| − | [[File: | + | [[File:web1.png|center|400px]]</div><br /> |
| − | |||
<!-----------------Classification----------------------> | <!-----------------Classification----------------------> | ||
| Line 48: | Line 46: | ||
<div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Classification</div> | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Classification</div> | ||
<div style="padding:0.5em 0.5em;"> | <div style="padding:0.5em 0.5em;"> | ||
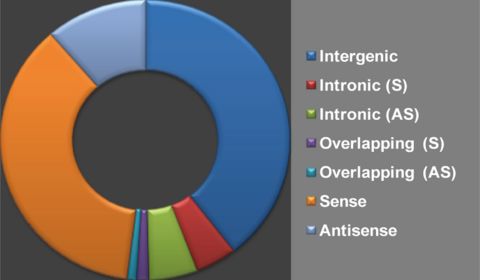
| − | + | The classification of lncRNA based on genomic location is now the most feasible classification method and is suggested to be of great biological significance (Ma etal. 2013, RNA Biology). <br /> | |
| − | We classified lncRNAs into | + | We classified lncRNAs into seven groups ('''Intergenic''', '''Intronic (S)''','''Intronic (AS)''','''Overlapping (S)''','''Overlapping (AS)''', '''Sense''' and '''Antisense''') based on their genomic location in respect to protein-coding genes. [[Help:Contents#Database content|'''''Help''''']]<br /> |
| − | [[File: | + | [[File:web2.png|center|480px]] |
</div> | </div> | ||
|} | |} | ||
Revision as of 08:38, 22 July 2014
|
Welcome to LncRNAWiki
52,616 human long non-coding RNAs inside
|
|
Introduction
LncRNAWiki is a wiki-based database for management of human lncRNAs, aiming to harness collective intelligence to collect, edit, and annotate information about human lncRNAs. This database is a publicly editable and open-content platform for community curation of human lncRNAs, bearing the potential to become a comprehensive and up-to-date knowledge base for human lncRNAs.
A featured lncRNA, ENST00000534336.1, that is overexpressed in many kinds of cancers and closely associated with tumor growth and metastasis, is community-curated. |
|
Data sources
|
Classification
The classification of lncRNA based on genomic location is now the most feasible classification method and is suggested to be of great biological significance (Ma etal. 2013, RNA Biology). |
|
Publications
|
About Us
Our group works in the field of Computational Biology and Bioinformatics (CBB), currently with a particular focus on building biological knowledge wikis in aid of community curation of massive biological knowledge.
|
Visitor Statistics
|