NONHSAT076153
PCGEM1 (Prostate cancer gene expression marker 1) is a long non-coding RNA (lncRNA) overexpressed in prostate cancer (PCa) cells that promotes PCa initiation and progression and protects against chemotherapy-induced apoptosis [1]. PCGEM1 has been characterised as a high-risk PCa marker and a potential biomarker for neoplasms responsive to chemoprevention by phytosterols [2].
Contents
Annotated Information
Name
PCGEM1: Prostate-specific transcript, LINC00071 (HGNC)
Characteristics
PCGEM1 is located at human chromosome 2q32.3 (HGNC), consists of three exons that comprehends 1603 bp (GenBank) [3].
Function
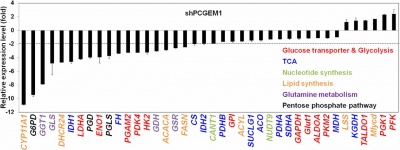
PCGEM1 regulates prostate cancer cell growth and tumor metabolism [4][1][2]. PCGEM1 regulates metabolism at a transcriptional level that affects multiple metabolic pathways, including glucose and glutamine metabolism, the pentose phosphate pathway, nucleotide and fatty acid biosynthesis, and the tricarboxylic acid cycle. The PCGEM1-mediated gene regulation takes place in part through AR activation, but predominantly through c-Myc activation, regardless of hormone or AR status. PCGEM1 binds directly to target promoters, physically interacts with c-Myc, promotes chromatin recruitment of c-Myc, and enhances its transactivation activity [4].
Over-expression of PCGEM1 leads to inhibition of apoptosis induced by doxorubicin [5]. PCGEM1 and PRNCR1 interact with the androgen receptor (AR) bound at DNA-enhancer regions in a ligand-dependent fashion and facilitate the chromosomal looping between AR-bound enhancers and the promoter sequences of androgen-responsive genes [2].
Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells and nu/nu PCa tumor growth. Both downregulation of a tumor-promoting long noncoding RNA PCGEM1 or overexpression of the tumor suppressor miR-145 reduced the proliferation and invasive capacity of prostate cancer cells in vitro and in vivo [1].
Disease
prostate cancer
Expression
PCGEM1, was expressed exclusively in human prostate tissue, was dramatically upregulated in PCa tissues compared with normal prostate tissues. Prostate tissue-specific and prostate cancer-associated (Srikantan 2000). Cholesterols upregulate the expression of PCGEM1 even in androgen-insensitive prostate cancer cell lines while phytosterols reverse this effect (Ifere 2009). PCGEM1 overexpression in LNCaP and in NIH3T3 cells promotes cell proliferation and a dramatic increase in colony formation, suggesting a biological role of PCGEM1 in cell growth regulation (Petrovics 2004). Low specificity as a biomarker for prostate cancer (14%) (Bialkowska-Hobrzanska 2006).
| Experiment | Forward primer | Reverse primer |
|---|---|---|
| Quantitative PCR | 5′-TGCCTCAGCCTCCCAAGTAAC-3′ | 5′-GGCCAAAATAAAACCAAACAT-3′[6] |
| siRNA | 5′-GCCCUACCUAUGAUUUCAUAU-3′ | 5′-AUAUGAAAUCAUAGGUAGGGC-3′[1] |
| qRT-PCR | 5′-CACGTGGAGGACTAAGGGTA-3′ | 5′-TTGCAACAAGGGCATTTCAG-3′[1] |
Regulation
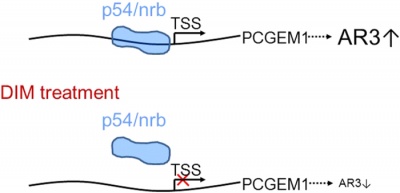
p54/nrb interact with the PCGEM1 promoter and regulate PCGEM1 [7].
Allelic Information and Variation
PCGEM1 polymorphisms may contribute to PCa risk in Chinese men. Men carrying single nucleotide polymorphisms (SNPs) of PCGEM1, i.e. rs6434568 AC and rs16834898 AC, had a lower PCa risk in comparison to the ones harboring CC and AA genotypes, respectively [8].
Labs working on this lncRNA
- Howard Hughes Medical Institute, Department of Medicine, University of California San Diego, La Jolla, California 92093, USA.
- Department of Molecular and Cellular Oncology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
- Department of Urology, School of Medicine, University of California Davis, Sacramento, California 95817, USA.
- Graduate Program, Kellogg School of Science and Technology, The Scripps Research Institute, La Jolla, California 92037, USA.
- Bioinformatics and System Biology Program, Department of Bioengineering, University of California San Diego, La Jolla, California 92093, USA.
- Neurosciences Graduate Program, Department of Biological Sciences, University of California San Diego, La Jolla, California 92093, USA.
- State Key Laboratory of Reproductive Medicine, Institute of Toxicology, Nanjing Medical University, Nanjing, China.[8]
- Department of Environmental Genomics, Jiangsu Key Laboratory of Cancer Biomarkers, Prevention and Treatment, Cancer Center, Nanjing Medical University, Nanjing, China.[8]
- Department of Genetic Toxicology, Key Laboratory of Modern Toxicology of Ministry of Education, School of Public Health, Nanjing Medical University, Nanjing, China.[8]
- Department of Urology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China.[8]
- Department of Environmental Genomics, School of Public Health, Nanjing Medical University, 818 East Tianyuan Road, Nanjing, Jiangsu 211166, China. [8]
- Department of Laboratory, Central Hospital of Panyu District, 8 Fuyu Dong Road, shiqiao, Guangzhou, Guangdong 511400, P R China.[1]
- The Second Affiliated Hospital of Guangzhou Medical University, Guangzhou 510620, China.[1]
- Department of Pharmacology and Toxicology, Cancer Institute, University of Mississippi Medical Center, Jackson, MS, USA.[7]
- Department of Radiation Oncology, University of Mississippi Medical Center, Jackson, MS, USA.[7]
- Department of Biochemistry, Cancer Institute, University of Mississippi Medical Center, Jackson, MS, USA.[7]
- Department of Radiation Oncology, Duke University Medical Center, Durham, NC, USA.[7]
- System Biosciences, Mountain View, CA, USA.[7]
- Department of Pulmonary Medicine, Tongji Hospital, Tongji University, Shanghai, China.[7]
- Department of Pathology, University of Mississippi Medical Center, Jackson, MS, USA.[7]
- College of Life Sciences, Zhejiang Sci-Tech University, Hangzhou, China.[7]
- Department of Biochemistry and Molecular Medicine, Comprehensive Cancer Center, University of California at Davis, Sacramento, CA 95817.[4]
- Institute of Molecular and Genomic Medicine, National Health Research Institutes, Zhunan Town, Miaoli County 350, Taiwan.[4]
- Department of Surgery, Center for Prostate Disease Research, Uniformed Services University of the Health Sciences, Bethesda, MD 20814-4799.[4]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 He J H, Zhang J, Han Z P, et al. Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells[J]. Journal of Experimental & Clinical Cancer Research. 2014, 33(1):72.
- ↑ 2.0 2.1 2.2 Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T & Calin GA. Long Noncoding RNA in Prostate, Bladder, and Kidney Cancer[J]. European Urology. 2014, 65(6):1140-1151.
- ↑ Srikantan V, Zou Z, Petrovics G, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer[J]. Proceedings of the National Academy of Sciences. 2000, 97(22): 12216-12221.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Hung C L, Wang L Y, Yu Y L, et al. A long noncoding RNA connects c-Myc to tumor metabolism[J]. Proceedings of the National Academy of Sciences. 2014, 111(52):18697-18702.
- ↑ Fu X, Ravindranath L, Tran N, Petrovics G & Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1[J]. DNA and cell biology. 2006, 25(3):135-141.
- ↑ Srikantan V, Zou Z, Petrovics G, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer[J]. Proceedings of the National Academy of Sciences. 2000, 97(22):12216-12221.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 7.7 7.8 7.9 Ho T T, Huang J, Zhou N, et al. Regulation of PCGEM1 by p54/nrb in prostate cancer[J]. Scientific reports, 2016, 6: 34529.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 Xue Y, Wang M, Kang M, et al. Association between lncrna PCGEM1 polymorphisms and prostate cancer risk[J]. Prostate cancer and prostatic diseases. 2013, 16(2):139.
Basic Information
| Transcript ID |
NONHSAT076153 |
| Source |
NONCODE4.0 |
| Same with |
, |
| Classification |
intergenic |
| Length |
1603 nt |
| Genomic location |
chr2+:193614571..193641625 |
| Exon number |
3 |
| Exons |
193614571..193614754,193615515..193615598,193640304..193641625 |
| Genome context |
|
| Sequence |
>gi |