Lnc-CDKN1A-1:1
PANDAR: "promoter of CDKN1A antisense DNA damage activated RNA", a long non coding RNA that regulates the response to DNA damage.
Contents
Annotated Information
Name
PANDAR:promoter of CDKN1A antisense DNA damage activated RNA (HGNC:44048)
Alias
PANDA: p21 associated ncRNA DNA damage activated (HGNC:44048)
Characteristics
The PANDAR was located approximately 5 kilobases upstream of the CDKN1A transcription start site and was induced upon DNA damage. [1]
Sequence runs antisense through a processed LAP3 pseudogene and contains a SINE.
Function
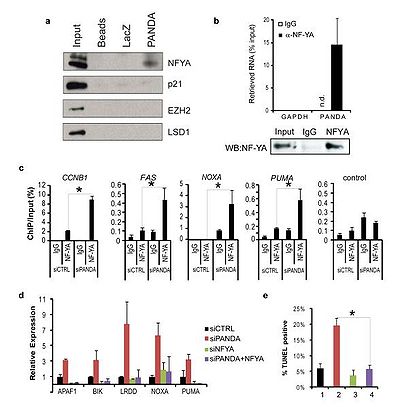
PANDA interacts with the transcription factor NF-YA to limit expression of pro-apoptotic genes; PANDA depletion markedly sensitized human fibroblasts to apoptosis by doxorubicin.[1]
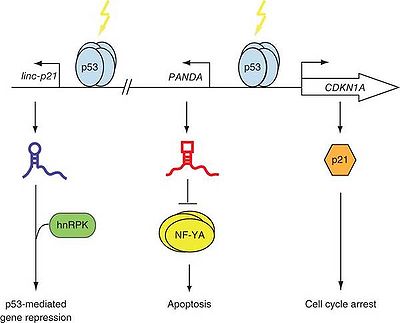
DNA damage activates p53-mediated transcription at CDKN1A and PANDA that functions synergistically to mediate cell cycle arrest and survival.[1]
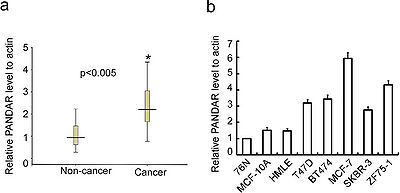
PANDAR could function as a tumor-promoting gene in breast cancer by regulating G1/S transition. [2]
Regulation
During DNA damage, PANDAR is induced by p53. [1]
Expression
PANDA interacts with the transcription factor NF-YA to limit expression of pro-apoptotic genes; PANDA depletion markedly sensitized human fibroblasts to apoptosis by doxorubicin.[1] One of five lncRNAs surrounding the transcription start site of the cell cycle gene CDKN1A that, along with the CDKN1A protein coding gene, were induced in human fetal lung fibroblasts upon DNA damage by doxorubicin.[1]
Expression of PANDA and CDKN1A were positively regulated by p53.[1]
Expression of neither CDKN1A itself nor TP53 was affected by PANDA depletion, suggesting that PANDA is a P53 effector that acts independently of p21-CDKN1A.[1]
PANDAR is up-regulated in breast cancer clinical samples as well as cell lines.[2]
PANDAR enhanced the binding of Bim1 complex to p16INK4A promoter and suppressed p16INK4A expression.[2]
| Primers | Fwd | Rev |
|---|---|---|
| Primers used in RACE | 5'-CAGAACTTGGCATGATGGAG-3'
5'-TGCACACATTTAACCCGAAG-3'[1] |
5'-TGATATGAAACTCGGTTTACTACTAGC-3'
5'-CCCCAAAGCTACATCTATGACA-3' 5'-CGTCTCCATCAT GCCAAGTT-3' 5'-CATAGAGCTTCACCGACATAGC-3'[1] |
| RT-PCR primers for PANDAR | 5'-TGCACACATTTAACCCGAAG-3' | 5'-CCCCAAAGCTACATCTATGACA-3'[1][2] |
| siRNAs for PANDAR | 5'-AAUGUGUGCACGUAACAGAUU-3' | 5'-GAGAUUUGCAGCAGACACAUU-3'[1] |
| 5'-GGGCAUGUUUUCACAGAGGUU-3' | 5'-GAGAUUUGCAGCAGACACAUU-3'[1] | |
| 5'-AAUGUGUGCACGUAACAGAUU-3' | 5'-GGGCAUGUUUUCACAGAGGUU-3'[1] | |
| ChIP assay | 5'-CCCATTTTCCTATCTGC-3' | 5'-CTAGTTCAAAGGATTCC-3'[2] |
Diseases
- Breast cancer [2]
Labs working on this lncRNA
- Program in Epithelial Biology, Stanford University School of Medicine, Stanford, California, USA.[1]
- Nanchang Key Laboratory of Cancer Pathogenesis and Translational Research, Center Laboratory, the Third Affiliated Hospital, Nanchang University, Nanchang, P.R. China.[2]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters[J]. Nature genetics. 2011,43(7):621-9.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Sang Y, Tang J, Li S, Li L, Tang X, Cheng C, et al. LncRNA PANDAR regulates the G1/S transition of breast cancer cells by suppressing p16(INK4A) expression[J]. Scientific reports. 2016,6:22366.
Basic Information
| Transcript ID |
lnc-CDKN1A-1:1 |
| Source |
LNCipedia2.1 |
| Same with |
, |
| Classification |
intergenic |
| Length |
1506 nt |
| Genomic location |
chr6+:36641397..36642903 |
| Exon number |
, |
| Exons |
, |
| Genome context |
|
| Sequence |
000001 ACGAATTCTT TCAGGAATGC CGCAGATGTA CATGCTCCCG CAGATCTATA TTTTCCAATG TTGTTAACAT CAGCCAGCTG 000080
000081 GCAATCTACA ACCTGTCTTG TACAATGTTT GAAGAGAGGC ATCCTCCAGA CACGGTCCCC TGTTTCAATG CTGGCCTCGA 000160 000161 AGAGCTTGTT CCAGAGCCAG GATGAATTGG TAAAGACCCC AGTGGCACCT GACCCCAAAG CTACATCTAT GACACCTGTT 000240 000241 AAGGTGGTGG CATTGAGGAT GACCTTCGGG TTAAATGTGT GCACGTAACA GAGCGCATCA GCCAGTATGA GCCTCCCCTC 000320 000321 AGCATCAGTG TTACCAACCT GGATGGTCTT CCTGTTCCTG GCTCTAACAA CATCCCCCAG CTTGTTGGCC TTGCCGCTGG 000400 000401 GCATGTTTTC ACAGAGGGGC CAGACCTATA ATATTAATGG GCAAACTGAG ATTTGCAGCA GACACAATGG CTGAGCATAT 000480 000481 AGTTGTAGCT CCTCCCATGT CGGCCCTCAT GAGGTCCATA TTTGCAGAAG CCTTGATGGA GATACCACCA CTGTCAAAGG 000560 000561 TAATTCCTTT CCCAACAAAC AAGGGGTGGT TTGTCTGCAT TGGGGCTGCC TATGTAGTGA ATTTCCAAGA AGACTGAGGG 000640 000641 CTCGTCAGAT CCTTTGGCCA CACTGAGGAA TGATCCCATT GCCTGTTCCT CAATCCAAGA CCTGGGTCTG ATATGAAACT 000720 000721 CGGTTTACTA CTAGCGCTTT TGAGATTCTT CTCAATAATT TCGGCAAATC TGGTTGGCAT CATCTCGCTG GCTGGCGTCT 000800 000801 CCATCATGCC AAGTTCTGCC CAGAAGCAAA CAGGACTCCT TTCTGCCAGG CCTCCTGATC CCCAGTTCCA TAGAGCTTCA 000880 000881 CCGACATAGC CATCTTCTTT TTTTGCTTTA GGTCATCGTA TTCATAGAGA CCAAGCACCG CGCCCTCCTC AGCAGCCTGA 000960 000961 GCATCTCTAC AGGGATCCAC CTCCACGGAA GAGAGCTCCA GGTCTTGAAT CTGCCTGCAT CCTGCTGCAA CAGCAGCTCT 001040 001041 GATGTTTTCT TTGCCTTCCT GCCAGTTTTC CTGTTCGTCG ATTCTGGCTG CCTTTTTGCC GAGGCCAACT AGCACCACGC 001120 001121 TGGGGAAGTC CTGATGCAGA CCATAAAAGT TTCGAGTCTT GCCTGCCTTC AGAGGTGGTC CAGATATGTT CAAAGTCTCT 001200 001201 CTCAGCTTTC CAGCTATCAA TTTATCAAGA TTCTCTCCTG CACTTGTGAA CTGTGGCACA TCATCTTCTT TTTCTTTGGA 001280 001281 ATAGATTCCT AAAACAAGGC CCTTCGTCAT GTCTGCAGCG GAGAGACCCC GGCTCCCGAA ACATCTCACG GCCAGACGTC 001360 001361 AGACGATGAC TCGCCCCACC CAATGTTTGT ATTTTAGTAG AGAGAGGGTT TCTCTATGTT GGTCAGGCTG GTCTCAAACC 001440 001441 TCGACCTCAG TTGATCTGTC TGCCTCGGTC TCCCAAAGTG CTGAGATTAC AGGCGTGAGC CACTGC |