FLVCR1-AS1
Contents
Annotated Information
Name
Approved symbol: FLVCR1-DT
Approved name: FLVCR1 divergent transcript
HGNC ID: HGNC:39077
Previous symbols: NCRNA00292; FLVCR1-AS1
Previous names: non-protein coding RNA 292; FLVCR1 antisense RNA 1 (non-protein coding); FLVCR1 antisense RNA 1; FLVCR1 antisense RNA 1 (head to head)
Alias symbols: LQK1
RefSeq ID: NR_027285
Ensembl ID: ENSG00000198468
GenBank accession number: AY030238 and AY030239
LncBook ID: https://bigd.big.ac.cn/lncbook/transcript?transid=HSALNT0288978 HSALNT0288978]
Chromosome
1q32.3
Characteristics
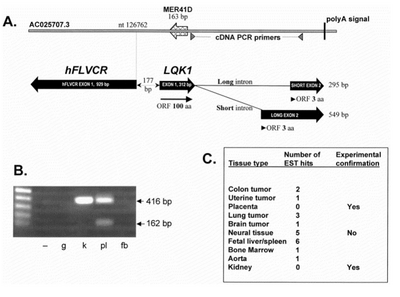
FLVCR1-AS1 is a 2-exon gene and it appears to have arisen after duplication of the hFLVCR locus.[1] FLVCR1-AS1 has two transcripts and both exon 2 isoforms share the same polyA signal.(Figure A )
Expression
FLVCR1-AS1 is expressed in a wide variety of tissues, including colon tumor, uterine tumor, placenta, lung tumor, brain tumor, neural tissue, fatal liver/spleen, bone marrow, aorta and kiney.(Figure C ) [1]
lncRNA FLVCR1‐AS1 was extremely up‐regulated in HCC tissues and cell lines. [2]
Function
Both splice forms of the FLVCR1-AS1 cDNA encode identical 103 aa ORFs that include a 163 nt MER41D interspersed repeat. [1]
FLVCR1‐AS1 acted as a competitive endogenous RNAs to sponge miR‐513c which targeted the mRNA of MET for degradation. By directly sponging miR‐513c, FLVCR1‐AS1 increased MET expression in HCC, and then promoted HCC progression.[2]
Disease
hepatocellular carcinoma [2]
Labs working on this lncRNA
- Division of Medical Genetics, Department of Medicine, University of Washington, Seattle, WA 98195-7710, USA. [1]
- Department of Pancreato-Biliary, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong Province, P.R. China.[2]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Lipovich L, Hughes AL, King MC, Abkowitz JL, Quigley JG. Genomic structure and evolutionary context of the human feline leukemia virus subgroup C receptor (hFLVCR) gene: evidence for block duplications and de novo gene formation within duplicons of the hFLVCR locus. Gene. 2002 Mar 20;286(2):203-13.
- ↑ 2.0 2.1 2.2 2.3 Zhang K, Zhao Z, Yu J, et al. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma[J]. J Cell Biochem, 2018, 119: 6045-6056.
Sequence
>gi|224591396|ref|NR_027285.1| Homo sapiens FLVCR1 antisense RNA 1 (head to head) (FLVCR1-AS1), transcript variant 1, long non-coding RNA
000081 GGGTCTCCTG CACCTGAGGG TGGGCGGGAG GCGGCCCCGC GGGTTGTGTT GCTGGAAAAA GGGGTCTCGA TCCAGACCCC 000160
000161 AAGAGAGGGT TCTTGGGTCT ACTTCAGGGA AGAATTGGAG GCGAGTCACA GAGCGCAGTG AAGGAAGCAA GTTTATTGGA 000240
000241 ATCTACTCCG TTAGAGAGTG CAAGTCCTCA GACTGCAGGA GGAGGAACTC CCGTCCTTCG GTAGTGTCAC TACTTAGAGG 000320
000321 CAGCTGTGAG GAGCTGTGAT TAATCGTGGA ATGTGCTGAT GTGCCCACTA AAGATGCCTG GTAAGCAGTG GCTCCTCTAA 000400
000401 CAAGTGGGCA GCTTGGAACT GGGAACGAGA GAGCCACCGA AACAGAATGA GGGAGACAAA GAAGTGGGCA TCACCCTTAA 000480
000481 CTCCACAAGG GCTTCCCAAG GGCGCTGTAC CGTACGCAGG CCCAGGACGA ACTTTATTTC TCGCCCCAGC ATCGCTGTCC 000560
000561 TTGTCGGTGA GACCCTGGCT TTAGGGCAGA CAGGACCACG TTTCATAAGT TCATGCTGTC CCAGCAGAGG AATAACGCCA 000640
000641 GAAAGTGTTC CAGTACAACC AGAGAAAGAG AGTCCATGAG AAATCTGCCC TTGTGAAGTT GGAATCCCCT CAACCTCACC 000720
000721 CCGCTGACTT GAATGAAGCG ACTGAGACGG GCTATGATGG AGCAGATCCG GTTGCTCGAC CTTCTCCCTT GCACCAACAC 000800
000801 ATGTAGTTAA TAGTTACTGG ACATGCATAT TCAGTGGGTT CCAGGTACCA AACTTGTATT GAATGGTATG TGCCAGACAC 000880
000881 CGTCTTGAGA TCTGGAGAAT AAAAAAATAA AAAAATAAAA ACGAGACATC CAGAGCACAG TA