ZFPM2-AS1
Novel lncRNA, ZFPM2-AS1 has a critical role in gastric carcinogenesis.
Contents
Annotated Information
Approved Name
Approved symbol:ZFPM2-AS1
Approved name:ZFPM2 antisense RNA 1
HGNC ID:50698
Alias symbol:SCAT3
RefSeq ID:NR_125796
Characteristics
ZFPM2-AS1 is located on chromosome 8q23, and three transcripts of the ZFPM2-AS1 gene are annotated in the National Center for Biotechnology Information database. However, the expression pattern and biological function of ZFPM2-AS1 in gastric cancer have yet to be reported [1].
Function
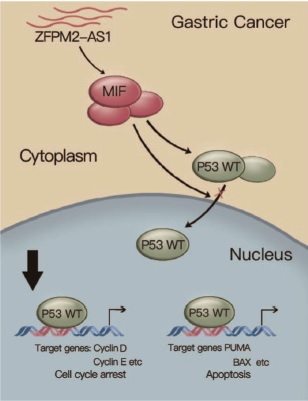
DNA methylation analyses of S-phase lncRNAs reveals that DNA methylation may underlie the differential expression of S-phase lncRNAs between tumor and normal tissue pairs. ZFPM2-AS1 is one such example which is upregulated in six types of cancers and this is consistent with its promoter’s hypomethylation in several cancers [2]. ZFPM2-AS1 expression promotes proliferation and suppresses apoptosis of gastric cancer cells in vitro and promotes tumor growth in vivo. This effect is associated with attenuated nuclear translocation of p53. Mechanistic experiments demonstrate that tumor-activated ZFPM2-AS1 could bind to and protect the degradation of macrophage migration inhibitory factor (MIF), a potent destabilizer of p53. Knockdown of MIF expression diminishes ZFPM2-AS1’s impact on p53 expression in gastric cancer cells [1].
Regulation
Please input information here.
Diseases
- Gastric carcinogenesis [1]
Expression
ZFPM2-AS1 expression is reported to be higher in gastric tumors than in normal gastric tissue. Also, increased ZFPM2-AS1 expression in gastric cancer specimens is associated with tumor size, depth of tumor invasion, differentiation grade, and TNM stage [1].
| Experiment | Forward primer | Reverse primer |
|---|---|---|
| qRT-PCR | 5'-CAATGGGACTAAGCCAGGCA- 3' | 5'-GGGCTCCACCAACAACCATA- 3'[1] |
Labs working on this lncRNA
- Department of Medical Biochemistry and Cell Biology, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, 40530 Sweden. [2]
- Department of Pediatrics, Institution for Clinical Sciences, Sahlgrenska Academy, University of Gothenburg, Gothenburg, 40530 Sweden. [2]
- Department of Gastroenterology, Changhai Hospital, Second Military Medical University, Shanghai, People's Republic of China. [1]
- Department of Gastroenterology, Hepatology & Nutrition, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA. [1]
- Department of Laboratory Medicine, Huashan Hospital Shanghai Medical College, Fudan University, Shanghai, People's Republic of China. [1]
- Department of Gastroenterology, Changhai Hospital, Second Military Medical University, Shanghai, People's Republic of China. [1]
- Department of Laboratory Medicine, Huashan Hospital Shanghai Medical College, Fudan University, Shanghai, People's Republic of China. [1]
- Department of Gastroenterology, Hepatology & Nutrition, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA. [1]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 Kong F, Deng X, Kong X, Du Y, Li L, Zhu H, Wang Y, Xie D, Guha S, Li Z, Guan M. ZFPM2-AS1, a novel lncRNA, attenuates the p53 pathway and promotes gastric carcinogenesis by stabilizing MIF. Oncogene. 2018, 37(45):5982.
- ↑ 2.0 2.1 2.2 Ali MM, Akhade VS, Kosalai ST, Subhash S, Statello L, Meryet-Figuiere M, Abrahamsson J, Mondal T, Kanduri C. PAN-cancer analysis of S-phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nature communications. 2018, 9(1):883.
Sequence
>gi|102723356|ref|NR_125796.1| Homo sapiens ZFPM2 antisense RNA 1 (ZFPM2-AS1), long non-coding RNA
000081 TTCCTGTACA CATCTTATCA ACAAAGAATT TGAACCTCAA AGAGTTCTGG TGCTGGTGAT TATTAAGTTC TTCACACTTC 000160
000161 TCTCATAGTG GAGCTTAAGA TTCTCACTGT GTTACCTAGG CTGCAGTGCA GTGGTGCGAT CTCAACTCAC TGCAACCTCT 000240
000241 ACCTCCTGGG TTCAAGCAAT TCTCCTGCCT CAGCCTCCCA AGTAGCTGGG ATTACAGAGC TCATGGTTTT TAAACAACTT 000320
000321 GTGGTAAAAA TCTGGTGGAA AAGATAACAA CAGTATATGA TATGGCAGAG TTGCACAGAA GAATCAGAAC ATTGTTTTAG 000400
000401 AGAAACGTTG GGCAATTAAT TAAGCCAGCT GATTAAGTTT TAAAGAAGCT AAACCATATC ATGTCGAAAT CAGAGGAGCG 000480
000481 ATGAAAGTGT GAGTGGTGGT ATTTCTGCTG TTCTCCCCCC TGAAGGCTTC TGCGATTAGC TTCTATGCCT TCCTTCCCTT 000560
000561 ATGGGCATTC AGATCCAAAC TACTTGCAAC TGTAGACAAG GAGGAAGATG GAACTTGGAG AAGACATACA GGCTGAAGAA 000640
000641 AAAAAAATAT TTTGGAAGTG ATCAGCACAT TCTAATTGAC CATGGATGCT CTCTGCATTC ATGTGGTAAC CCAGGGAGAG 000720
000721 TATGGAGTGA AAAGAGGATC ACCAAAGAAA CTTGGGGAAT TACACACTAA TAGTGAAGAA CAGGTGGCAC CTGAAATCAC 000800
000801 AGAAAATTTA TTTGTCCTCA ATGCAAAGAT GAACTATGAA GAGCACTTTA TTGCGTTTGA AGAATATAGG AATGATGAGT 000880
000881 GAAATGGTGA AGGAAGTGGA TAAACAATGA TGTAACGACT GAGCGTCATC TTGCAACTGA ATAAGCCAGT GAGGAACTTG 000960
000961 GTAAATTTGT TGATTATATT TAGTTTTACA ATGTTTAATT CTTTATTATT T