ENST00000534336.1
MALAT-1 is overexpressed in many kinds of cancers and is closely associated with tumor growth and metastasis.
Contents
Annotated Information
Transcriptomic Nomeclature
N11QT0096001-SSCY-LMXXX01 Help
Name
Malat1: Metastasis-associated lung adenocarcinoma transcript 1
Neat2: Nuclear enriched abundant transcript 2
Characteristics
Malat1 gene is localized in the intergenic region of the genome. It is about 8kb in length and the transcript has only one exon [1][2]. The 3’-end of the transcript is a conserved tRNA-like sequence, which can be modified by RNase P and cleaved by RNase Z to yield another ncRNA (61nt), the cytoplasmic MALAT1-associated small cytoplasmic RNA (mascRNA) [3].
Malat1 is stable in human B cells (half-life: 16.5 h) [4] and Hela cells (half-life: about 7 h) [5], but is unstable in mouse 3T3 cells (half-life: 3 h) [4] and N2A cells ( half-life: 4 h) [4]. MALAT-1 is generally stable in cancer cells, with the half-life ranging from ~ 9 h to > 12 h in various cancer cells [6]. Xrn2, PM/Scl-75, PARN, and Mtr4, known nuclear RNases or RNA helicases, do not affect MALAT-1 degradation [6].
Cellular localization
MALAT-1 localizes predominately in the nucleus [2]. In G2/M cell cycle phase, MALAT-1 transcripts partially translocate from the nucleus into the cytoplasm [7]. RNPS1, SRm160, and IBP160 are found to contribute to the nuclear localization of MALAT-1 [8].
Function
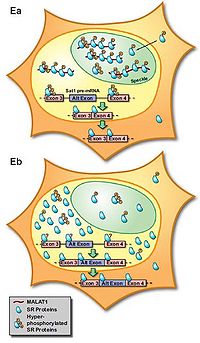
MALAT-1 localizes specifically in the SC35 splicing domains in the nucleus, suggesting its function in pre-mRNA metabolism or specific nuclear structures [2]. The later studies found that MALAT-1 could modulate mRNA alternative splicing via its interaction with the serine/arginine-rich (SR) family of nuclear phosphoproteins that are involved in the splicing machinery [9]. MALAT-1 regulates synapse formation by modulating the expression of genes involved in synapse formation and/or maintenance [10]. However, splicing alterations were not found after Malat1 ablation in mice [11]. Also, MALAT1 does not alter alternative splicing but actively regulates gene expression including a set of metastasis-associated genes in lung cancer cells [12]. These results indicate that alternative functions for MALAT-1 may exist.
MALAT-1 can function by participating in localization of important proteins, such as hnRNP C [7] and growth control genes [13]. MALAT-1 is found to regulate cell cycle progress in G2/M phase [7][9], G1/S phase [14], and G0/G1 phase [15]. In the G2/M phase, MALAT-1 interacts with hnRNP C to facilitate the cytoplasmic translocation of hnRNP C, leading to cell cycle progresion [7]. MALAT1 could interact with the demethylated form of CBX4 (chromobox homolog 4), and controls the relocalization of growth control genes between polycomb bodies and interchromatin granules [13]. MALAT1 could bind to Human PSF (hPSF) protein to release hPSF from a repressed proto-oncogene and activate transcription, driving transformation and tumorigenesis [16].
MALAT-1 regulates synaptogenesis [10]and is involved in the development of advanced invasive placentation [17]. Deregulation of MALAT-1 is found to be closely associated with the development of cancer. In vitro, it is found that MALAT-1 promotes epithelial–mesenchymal transition (EMT) of bladder cancer cells by activating Wnt signaling [18]. 3' end of MALAT-1 (6918 nt-8441 nt) is found to be important in colorectal cancer metastasis [19]. In lung adenocarcinoma cells, MALAT-1 may regulate cell motility through transcriptional and post-transcriptional regulation of motility related gene expression [20]. However, mechanisms of these functions are not clear.
Expression
MALAT-1 is ubiquity expressed in various normal tissues [1][2][3][10], but the expression levels are quite different among tissues [1][2][10].
MALAT-1 is over-expressed in many human carcinomas, including those of the breast, pancreas, lung, colon, prostate, and liver [21].
It is also found to be up-regulated in the cerebellum, hippocampus and brain stem of human alcoholics [22].
| Primer | Forward primer | Reverse primer |
|---|---|---|
| RT-PCR | 5'-AAAGCAAGGTCTCCCCACAAG-3' | 5'-GGTCTGTGCTAGATCAAAAGGCA-3'[23][24] |
| 5'-CTTCCCTAGGGGATTTCAGG-3' | 5'-GCCCACAGGAACAAGTCCTA-3'[15] | |
| 5'-GAATTGCGTCATTTAAAGCCTAGTT-3' | 5'-GTTTCATCCTACCACTCCCAATTAAT-3'[25] | |
| 5'-cggaagtaattcaagatcaagag-3' | 5'-actgaatccacttctgtgtagc-3'[16] | |
| cDNA amplication | 5'-GTAGGGCCCTCCATGGCGATTTGCCTTGTGAGCAC-3' | 5'-GAGCTCGAGGTCCTGAAGACAGATTAGTAGTCAAAGC-3'[6] |
| Northern blot | 5'-GGCAGGAGAGACAACAAAGC-3' | 5'-CTCGACACCATCGTTACCT-3'[2] |
Regulation
In breast cancer cells, high concentration E2 treatment largely decreases MALAT-1 RNA level in an ERa independent way [26].
Disruption of p53 appears to play an important role in the up-regulation of MALAT-1 [27].
CREB (cyclic AMP-responsive element binding) transcription factor is found to bind to the defined proximal promoter of the MALAT1 gene, leading to the up-regulation of MALAT1 [28].
Diseases
MALAT-1 was first identified as a prognostic marker for metastasis and patient survival in non-small cell lung cancer (NSCLC) [29]. It is found to be overexpressed in various tumors and cancer cell lines, including lung cancer [1][21][29], endometrial stromal sarcoma of the uterus [30], hepatocellular carcinomas [21][31], breast cancer [21][32], pancreas cancer [21], colon cancer [21], prostate cancer [15][21], melanoma [23], bladder cancer [18]. Overexpression of MALAT-1 in cancer cells is closely associated with tumor growth and metastasis [12][20][23][29].
There is no significant difference in MALAT-1 lncRNA levels in normal pituitary tissues, invasive NFPAs (non-functioning pituitary adenomas), and non-invasive NFPAs, and no significant association between MALAT-1 expression and patient clinicopathological characteristics [24].
Evolution
MALAT-1 is highly conserved across mammals [10]. However, sequence conservation is limited in vertebrates. Sequence similarity between zebrafish and mammalian MALAT1 is restricted to the 3′ end, while the length of MALAT1 (~7 kb) along with the gene structure appeare to be roughly fixed in all vertebrates [14].
Associated components
- hnRNP C [7]
- CBX4 (chromobox homolog 4) [13]
- Human PSF (hPSF) protein [16]
You can also add sub-section(s) at will.
Labs working on this lncRNA
- Department of Medicine, University of Münster, Germany
- Howard Hughes Medical Institute, University of California, San Diego, School of Medicine, 9500 Gilman Drive, La Jolla, CA 92093-0648, USA
- Department of Urology, Shanghai Changhai Hospital, Second Military Medical University, Shanghai, People's Republic of China
- Institute of Molecular Medicine and State Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University, Beijing 100871, China
References
- ↑ 1.0 1.1 1.2 1.3 Ji, P., Diederichs, S., Wang, W., Boing, S., Metzger, R., Schneider, P.M., Tidow, N., Brandt, B., Buerger, H., Bulk, E. et al. (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene, 22, 8031-8041.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Hutchinson, J.N., Ensminger, A.W., Clemson, C.M., Lynch, C.R., Lawrence, J.B. and Chess, A. (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics, 8, 39.
- ↑ 3.0 3.1 Wilusz, J.E., Freier, S.M. and Spector, D.L. (2008) 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell, 135, 919-932.
- ↑ 4.0 4.1 4.2 Clark, M.B., Johnston, R.L., Inostroza-Ponta, M., Fox, A.H., Fortini, E., Moscato, P., Dinger, M.E. and Mattick, J.S. (2012) Genome-wide analysis of long noncoding RNA stability. Genome Res, 22, 885-898.
- ↑ Tani, H., Mizutani, R., Salam, K.A., Tano, K., Ijiri, K., Wakamatsu, A., Isogai, T., Suzuki, Y. and Akimitsu, N. (2012) Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res, 22, 947-956.
- ↑ 6.0 6.1 6.2 Tani, H., Nakamura, Y., Ijiri, K. and Akimitsu, N. (2010) Stability of MALAT-1, a nuclear long non-coding RNA in mammalian cells, varies in various cancer cells. Drug Discov Ther, 4, 235-239.
- ↑ 7.0 7.1 7.2 7.3 7.4 Yang, F., Yi, F., Han, X., Du, Q. and Liang, Z. (2013) MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett, 587, 3175-3181.
- ↑ Miyagawa, R., Tano, K., Mizuno, R., Nakamura, Y., Ijiri, K., Rakwal, R., Shibato, J., Masuo, Y., Mayeda, A., Hirose, T. et al. (2012) Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA, 18, 738-751.
- ↑ 9.0 9.1 9.2 Tripathi, V., Ellis, J.D., Shen, Z., Song, D.Y., Pan, Q., Watt, A.T., Freier, S.M., Bennett, C.F., Sharma, A., Bubulya, P.A. et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell, 39, 925-938.
- ↑ 10.0 10.1 10.2 10.3 10.4 Bernard, D., Prasanth, K.V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., Zhang, M.Q., Sedel, F., Jourdren, L., Coulpier, F. et al. (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J, 29, 3082-3093.
- ↑ Zhang, B., Arun, G., Mao, Y.S., Lazar, Z., Hung, G., Bhattacharjee, G., Xiao, X., Booth, C.J., Wu, J., Zhang, C. et al. (2012) The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep, 2, 111-123.
- ↑ 12.0 12.1 Gutschner, T., Hammerle, M., Eissmann, M., Hsu, J., Kim, Y., Hung, G., Revenko, A., Arun, G., Stentrup, M., Gross, M. et al. (2013) The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res, 73, 1180-1189.
- ↑ 13.0 13.1 13.2 Yang, L., Lin, C., Liu, W., Zhang, J., Ohgi, K.A., Grinstein, J.D., Dorrestein, P.C. and Rosenfeld, M.G. (2011) ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell, 147, 773-788.
- ↑ 14.0 14.1 Ulitsky, I., Shkumatava, A., Jan, C.H., Sive, H. and Bartel, D.P. (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell, 147, 1537-1550.
- ↑ 15.0 15.1 15.2 Ren, S., Liu, Y., Xu, W., Sun, Y., Lu, J., Wang, F., Wei, M., Shen, J., Hou, J., Gao, X. et al. (2013) Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol, 190, 2278-2287.
- ↑ 16.0 16.1 16.2 Li, L., Feng, T., Lian, Y., Zhang, G., Garen, A. and Song, X. (2009) Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A, 106, 12956-12961.
- ↑ Tseng, J.J., Hsieh, Y.T., Hsu, S.L. and Chou, M.M. (2009) Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod, 15, 725-731.
- ↑ 18.0 18.1 Ying, L., Chen, Q., Wang, Y., Zhou, Z., Huang, Y. and Qiu, F. (2012) Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst, 8, 2289-2294.
- ↑ Xu, C., Yang, M., Tian, J., Wang, X. and Li, Z. (2011) MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol, 39, 169-175.
- ↑ 20.0 20.1 Tano, K., Mizuno, R., Okada, T., Rakwal, R., Shibato, J., Masuo, Y., Ijiri, K. and Akimitsu, N. (2010) MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett, 584, 4575-4580.
- ↑ 21.0 21.1 21.2 21.3 21.4 21.5 21.6 Lin, R., Maeda, S., Liu, C., Karin, M. and Edgington, T.S. (2007) A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene, 26, 851-858.
- ↑ Kryger, R., Fan, L., Wilce, P.A. and Jaquet, V. (2012) MALAT-1, a non protein-coding RNA is upregulated in the cerebellum, hippocampus and brain stem of human alcoholics. Alcohol, 46, 629-634.
- ↑ 23.0 23.1 23.2 Tian, Y., Zhang, X., Hao, Y., Fang, Z. and He, Y. (2014) Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res.
- ↑ 24.0 24.1 Li, Z., Li, C., Liu, C., Yu, S. and Zhang, Y. (2014) Expression of the long non-coding RNAs MEG3, HOTAIR, and MALAT-1 in non-functioning pituitary adenomas and their relationship to tumor behavior. Pituitary.
- ↑ Guo, F., Li, Y., Liu, Y., Wang, J. and Li, G. (2010) Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai), 42, 224-229.
- ↑ Zhao, Z., Chen, C., Liu, Y. and Wu, C. (2014) 17beta-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem Biophys Res Commun, 445, 388-393.
- ↑ Jeffers, L.K., Duan, K., Ellies, L.G., Seaman, W.T., Burger-Calderon, R.A., Diatchenko, L.B. and Webster-Cyriaque, J. (2013) Correlation of transcription of MALAT-1, a novel noncoding RNA, with deregulated expression of tumor suppressor p53 in small DNA tumor virus models. J Cancer Ther, 4.
- ↑ Koshimizu, T.A., Fujiwara, Y., Sakai, N., Shibata, K. and Tsuchiya, H. (2010) Oxytocin stimulates expression of a noncoding RNA tumor marker in a human neuroblastoma cell line. Life Sci, 86, 455-460.
- ↑ 29.0 29.1 29.2 Schmidt, L.H., Spieker, T., Koschmieder, S., Schaffers, S., Humberg, J., Jungen, D., Bulk, E., Hascher, A., Wittmer, D., Marra, A. et al. (2011) The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol, 6, 1984-1992.
- ↑ Yamada, K., Kano, J., Tsunoda, H., Yoshikawa, H., Okubo, C., Ishiyama, T. and Noguchi, M. (2006) Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci, 97, 106-112.
- ↑ Lai, M.C., Yang, Z., Zhou, L., Zhu, Q.Q., Xie, H.Y., Zhang, F., Wu, L.M., Chen, L.M. and Zheng, S.S. (2011) Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol, 29, 1810-1816.
- ↑ Guffanti, A., Iacono, M., Pelucchi, P., Kim, N., Solda, G., Croft, L.J., Taft, R.J., Rizzi, E., Askarian-Amiri, M., Bonnal, R.J. et al. (2009) A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics, 10, 163.
Basic Information
| Transcript ID |
ENST00000534336.1 |
| Source |
Gencode19 |
| Same with |
lnc-SCYL1-1:2,NONHSAT022127 |
| Classification |
intergenic |
| Length |
8708 nt |
| Genomic location |
chr11+:65265233..65273940 |
| Exon number |
1 |
| Exons |
65265233..65273940 |
| Genome context |
|
| Sequence |
|