Difference between revisions of "Main Page"
| Line 32: | Line 32: | ||
</div> | </div> | ||
---- | ---- | ||
| + | |||
| + | |} | ||
<!--------------main part one----------------> | <!--------------main part one----------------> | ||
{|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | {|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | ||
| Line 38: | Line 40: | ||
|valign="top" style="border:1px solid #66B3FF; background-color:transparent; "| | |valign="top" style="border:1px solid #66B3FF; background-color:transparent; "| | ||
<div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Disease</div> | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Disease</div> | ||
| − | <div style="padding:0.5em 0.5em;">[[File:Statistics4.png| | + | <div style="padding:0.5em 0.5em;">[[File:Statistics4.png|left|900px]] |
| − | LncRNAWiki keeps frequent updates by community annotation of human lncRNAs and integration of newly identified lncRNAs with experimental evidence. In contrast to the previous version (released in 2014) that had 86 curated lncRNAs, the updated version has, a total of 719 lncRNAs that are community-curated (the detailed list is available at [[LncRNAWiki:Featured|Featured LncRNAs]]). | + | LncRNAWiki keeps frequent updates by community annotation of human lncRNAs and integration of newly identified lncRNAs with experimental evidence. |
| + | |||
| + | In contrast to the previous version (released in 2014) that had 86 curated lncRNAs, the updated version has, a total of 719 lncRNAs that are community-curated (the detailed list is available at [[LncRNAWiki:Featured|Featured LncRNAs]]). | ||
Among these 719 lncRNAs, 304 have been experimentally proved to be associated with cancer and other diseases ([[LncRNAWiki:Disease|Disease]]) | Among these 719 lncRNAs, 304 have been experimentally proved to be associated with cancer and other diseases ([[LncRNAWiki:Disease|Disease]]) | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| + | |||
<!--------------main part two----------------> | <!--------------main part two----------------> | ||
{|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | {|border="0" cellspacing="0" cellpadding="0" style="margin:1em 0; background:transparent; width:100%;" | ||
|- | |- | ||
| − | <!-----------Data | + | <!-----------Data source------------> |
|valign="top" style="border:1px solid #66B3FF; background-color:transparent; "| | |valign="top" style="border:1px solid #66B3FF; background-color:transparent; "| | ||
| − | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Data | + | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Data source</div> |
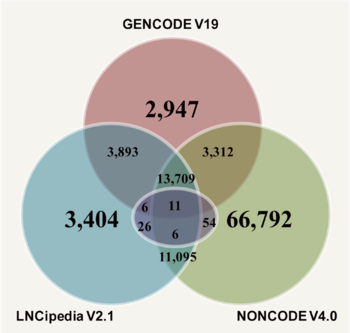
| − | <div style="padding:0.5em 0.5em;">[[File:web1a.png| | + | <div style="padding:0.5em 0.5em;">[[File:web1a.png|center|350px|caption]] |
| − | + | The majority of the lncRNA information was initially seeded with a subset of information from [http://www.gencodegenes.org/ GENCODE], [http://www.bioinfo.org/noncode/ NONCODE], [http://www.lncipedia.org/ LNCipedia], and [http://www.lncrnadb.org/ lncRNAdb](purple circle indicates lncRNAdb). | |
| − | |||
| − | The majority of the lncRNA information was initially seeded with a subset of information from [http://www.gencodegenes.org/ GENCODE], [http://www.bioinfo.org/noncode/ NONCODE], [http://www.lncipedia.org/ LNCipedia], and [http://www.lncrnadb.org/ lncRNAdb]. | ||
<br /></div> | <br /></div> | ||
<!-----------------Classification----------------------> | <!-----------------Classification----------------------> | ||
| − | + | |valign="top" style="border:1px solid #66B3FF; background-color:transparent; "| | |
| − | |valign="top" style="border:1px solid #66B3FF; background-color:transparent | ||
<div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Classification</div> | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Classification</div> | ||
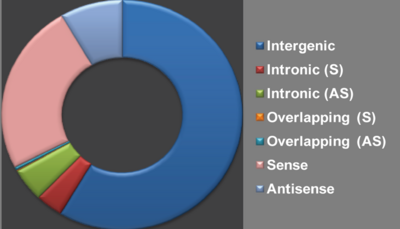
| − | <div style="padding:0.5em 0.5em;"> | + | <div style="padding:0.5em 0.5em;">[[File:web2a.png|center|400px]] |
| − | + | Based on genomic location, we classified lncRNAs into seven groups (Ma etal. 2013, RNA Biology) ('''Intergenic''', '''Intronic (S)''','''Intronic (AS)''','''Overlapping (S)''','''Overlapping (AS)''', '''Sense''' and '''Antisense''') based on their genomic location in respect to protein-coding genes. [[Help:Contents#Database content|''Help'']] | |
| − | + | <br /></div> | |
| + | |||
| + | <!-----------------Small Protein----------------------> | ||
| + | |valign="top" style="border:1px solid #66B3FF; background-color:transparent; "| | ||
| + | <div style="background:#ACD6FF; border-bottom:1px solid #66B3FF; padding:0.2em 0.5em; font-size:125%; font-weight:bold;">Small Protein</div> | ||
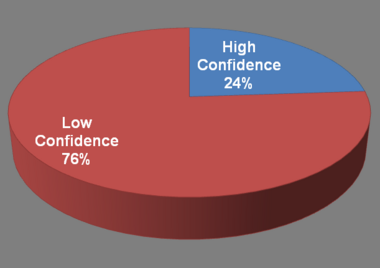
| + | <div style="padding:0.5em 0.5em;">[[File:SmallPRO-1.png|center|380px]] | ||
| + | We developed computational approaches for identification of small proteins in all the human lncRNAs in LncRNAWiki. As a result, we identified '''9,387''' lncRNAs potentially encoding small proteins and we further validated '''2,246''' such small proteins with high confidence by considering protein instability, secondary structure and transmembrane helix ([[LncRNAWiki:small proteins|lncRNAs and small proteins]], [[Help:Contents#Database content|help]]). | ||
| + | <br /></div> | ||
| − | |||
| − | |||
| − | |||
|} | |} | ||
Revision as of 09:06, 20 June 2017
|
Welcome to LncRNAWiki
105,824 human long non-coding RNAs inside
|
|
Introduction
LncRNAWiki is a wiki-based, publicly editable and open-content platform for community curation of human long non-coding RNAs (lncRNAs), viz., a community-curated lncRNA knowledgebase. Unlike conventional biological databases based on expert curation, lncRNAWiki harnesses collective intelligence to collect, edit and annotate information about lncRNAs, quantifies users' contributions in each annotated lncRNA and provides explicit authorship to encourage more participation from the whole scientific community. Imagine a world in which every single person on the planet has free access to the sum of all human knowledge. — Jimmy Wales, Founder of Wikipedia
|
|
Disease
LncRNAWiki keeps frequent updates by community annotation of human lncRNAs and integration of newly identified lncRNAs with experimental evidence. In contrast to the previous version (released in 2014) that had 86 curated lncRNAs, the updated version has, a total of 719 lncRNAs that are community-curated (the detailed list is available at Featured LncRNAs). Among these 719 lncRNAs, 304 have been experimentally proved to be associated with cancer and other diseases (Disease) |
|
Data source
|
Classification
Based on genomic location, we classified lncRNAs into seven groups (Ma etal. 2013, RNA Biology) (Intergenic, Intronic (S),Intronic (AS),Overlapping (S),Overlapping (AS), Sense and Antisense) based on their genomic location in respect to protein-coding genes. Help |
Small Protein
We developed computational approaches for identification of small proteins in all the human lncRNAs in LncRNAWiki. As a result, we identified 9,387 lncRNAs potentially encoding small proteins and we further validated 2,246 such small proteins with high confidence by considering protein instability, secondary structure and transmembrane helix (lncRNAs and small proteins, help). |
|
How to Cite
If you use any data or information in your work, please cite the following primary publication:
Related publication:
|
About Us
Our group works in the field of Computational Biology and Bioinformatics (CBB), currently with a particular focus on building biological knowledge wikis in aid of community curation of massive biological knowledge.
|
Visitor Statistics
|