Difference between revisions of "NONHSAT075638"
Chunlei Yu (talk | contribs) (→Transcriptomic Nomeclature) |
|||
| Line 21: | Line 21: | ||
===Diseases=== | ===Diseases=== | ||
metastatic cancer<ref name="ref2" /> | metastatic cancer<ref name="ref2" /> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
===Expression=== | ===Expression=== | ||
| Line 34: | Line 28: | ||
HOXD-AS1 is expressed in multiple tissues with the highest levels of expression in kidney, colon and testes and the lowest in liver, heart, pancreas and stomach. In general, HOXD-AS1 demonstrated highly correlated expression with its neighboring protein coding genes HOXD1 and HOXD3, thus implying that they are subject to the common regulatory mechanism<ref name="ref2" />. | HOXD-AS1 is expressed in multiple tissues with the highest levels of expression in kidney, colon and testes and the lowest in liver, heart, pancreas and stomach. In general, HOXD-AS1 demonstrated highly correlated expression with its neighboring protein coding genes HOXD1 and HOXD3, thus implying that they are subject to the common regulatory mechanism<ref name="ref2" />. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
===Evolution=== | ===Evolution=== | ||
| Line 50: | Line 33: | ||
More than 96% of this gene sequence is evolutionary conserved among hominids (H. sapiens, P. paniscus, P. troglodytes and P. abelii). At the same time, only 12% to 52% of HOXD-AS1sequence is shared with other primates (M. mulatta, N. leucogenys, C. jacchus and O. garnettii), comparable to 11% of dog (C. lupus) and 8-12% of ungulates (E. caballus, S. scrofa and B. taurus). Thus, a gap between HOXD-AS1 genomic sequence in apes and other monkeys is apparent<ref name="ref2" />. | More than 96% of this gene sequence is evolutionary conserved among hominids (H. sapiens, P. paniscus, P. troglodytes and P. abelii). At the same time, only 12% to 52% of HOXD-AS1sequence is shared with other primates (M. mulatta, N. leucogenys, C. jacchus and O. garnettii), comparable to 11% of dog (C. lupus) and 8-12% of ungulates (E. caballus, S. scrofa and B. taurus). Thus, a gap between HOXD-AS1 genomic sequence in apes and other monkeys is apparent<ref name="ref2" />. | ||
| − | |||
| − | |||
===Cellular Localization=== | ===Cellular Localization=== | ||
Revision as of 11:48, 30 June 2016
HAGLR, which is induced by RA and regulated via PI3K/Akt pathway, controls genes involved in RA signaling, angiogenesis and inflammation.
Contents
Annotated Information
Name
HAGLR:HOXD antisense growth-associated long non-coding RNA(HGNC nomenclature)
Mdgt[1]
"HOXD cluster antisense RNA 1", "HOXD cluster antisense RNA 1 (non-protein coding)", HOXD-AS1[2]
Characteristics
OXD-AS1 is evolutionary conserved among hominids and has all bona fide features of a gene[2].
Function

HOXD-AS1 is evolutionary conserved among hominids and has all bona fide features of a gene. HOXD-AS1 is a subject to morphogenic regulation, is activated by PI3K/Akt pathway and itself is involved in control of RA-induced cell differentiation. Knock-down experiments revealed that HOXD-AS1 controls expression levels of clinically significant protein-coding genes involved in angiogenesis and inflammation, the hallmarks of metastatic cancer[2].
Regulation
HOXD-AS1 is regulated by PI3K/Akt pathway[2].
Diseases
metastatic cancer[2]
Expression
Mdgt is highly expressed in testes, and more moderately in brain, kidney, colon and skeletal muscles[1].
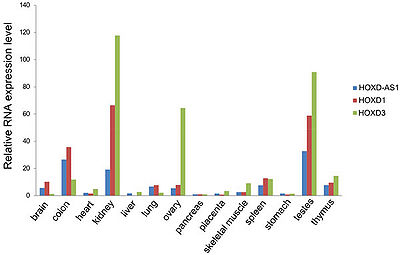
HOXD-AS1 is expressed in multiple tissues with the highest levels of expression in kidney, colon and testes and the lowest in liver, heart, pancreas and stomach. In general, HOXD-AS1 demonstrated highly correlated expression with its neighboring protein coding genes HOXD1 and HOXD3, thus implying that they are subject to the common regulatory mechanism[2].
Evolution
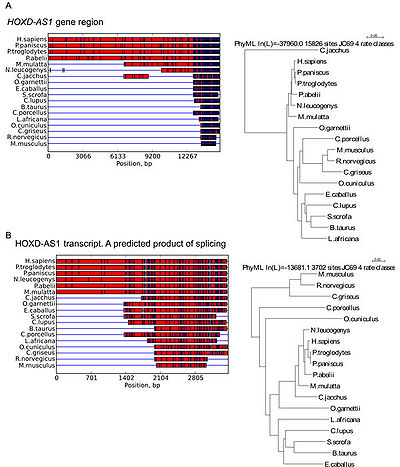
More than 96% of this gene sequence is evolutionary conserved among hominids (H. sapiens, P. paniscus, P. troglodytes and P. abelii). At the same time, only 12% to 52% of HOXD-AS1sequence is shared with other primates (M. mulatta, N. leucogenys, C. jacchus and O. garnettii), comparable to 11% of dog (C. lupus) and 8-12% of ungulates (E. caballus, S. scrofa and B. taurus). Thus, a gap between HOXD-AS1 genomic sequence in apes and other monkeys is apparent[2].
Cellular Localization
HOXD-AS1 is evenly distributed between nucleus and cytoplasm[2].
Labs working on this lncRNA
Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, United States[1].
Department of Genome and Gene Expression Data Analysis, Bioinformatics Institute, Agency for Science, Technology and Research (A*STAR), Matrix, 138671 Singapore[2].
References
- ↑ 1.0 1.1 1.2 Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development[J]. eLife. 2013,2:e01749.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 Yarmishyn AA, Batagov AO, Tan JZ, Sundaram GM, Sampath P, Kuznetsov VA, et al. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome[J]. BMC genomics. 2014,15 Suppl 9:S7.
Basic Information
| Transcript ID |
NONHSAT075638 |
| Source |
NONCODE4.0 |
| Same with |
HAGLR,Mdgt,HOXD-AS1 |
| Classification |
intergenic |
| Length |
3805 nt |
| Genomic location |
chr2-:177037924..177053686 |
| Exon number |
3 |
| Exons |
177037924..177041420,177042060..177042222,177053542..177053686 |
| Genome context |
|
| Sequence |
000001 AGCTGACGAA GGCGCCGTCG CCGTTGCCCA GAGGGAAGGC GGGCTGCAGA GCCACGGGCC GGGCGTCGGA GCGGCAGAAC 000080
000081 TTGGGTGCCA AGCTGAGCAC GTCGCCGCCG ACCCCGCCGC TGCTGCTGCA TGACACGTAC TCCAGATCTG GGGACCCTTC 000160 000161 ACCTGCCTCT ACTACTGCAA ATGATTGGAT ATTCGTCTGA CTTGGCTCTT CCCTAATGTG TGGAACTAAT TTTGTTCACT 000240 000241 CTGTTTCATG CTGGACCTGG CCTGAAAATG GGCCCTTATA TTGTCTTTGC ACTTTCCAAC TTGCCAAGTT TCTGAAAGAA 000320 000321 GGACCAAAGT AATGCCAAGA ACTCCCAGAT GAAGAGGACC TTCCTTAGAC CTATAACAGA TTCACTACTT GACTTCAGAG 000400 000401 GGAAGTTTCA GCTAATTCTC AGGAAAAGGT CAAAACAGGC CTGTGACTGT CCCTTGAATA CTGAAGGTAT AAACAAGCTT 000480 000481 CTGGCTCCTC CAGGTCAAGC AGAGGAACAC TTTTATTAGA ATGAAGGGTT ACTTCTTTGT TATTCAAACA CACACCCACA 000560 000561 CACATCCTCT GTAGTCTCCC TAATGAAAAA CCTCAACAGA TGGAGAGCCT GAGCTCCCAA GTCCGCAGGC TGAGAAGATG 000640 000641 CACTTCACTC AAGGGCCAGT GGGCTGGTAC AGACTAGGGA AGAGAAAAGG GACAAAGGCT GCTAGTATAG GTAATCACTA 000720 000721 TCTTTTAATA CTCCTAACAC ATAGAAATTC GAAAAATCTT GTTCAGCAAT GTTAGCAGCC TGTGAGCCTC GGAAACTAAT 000800 000801 TTGGCTGATG TGTCTCTGAT AGGGAGCTTG GGGCTGCACC AAGGGGAAAC ATTCTCCTCC TGTATTCTCT CCTCTCTCTC 000880 000881 GTTTGAAAGC CTGTACAAAG AAATGGTACA AATGGATGGT TAACTTAATG ATTCCATCAG AGGCAGATTT TTCTCTGTCA 000960 000961 GGTCCCTGGA TAAGGTGGAG GTCTGAAAGC CAGGAGACAG AATATGTACT TAATCGCCCT TTCTGACCTG CTTACCAGTG 001040 001041 ACCAGGACTA CTCTCCAGGC CTTTCTTCTG CCACTTCCCC ATTCTTCCTG GCTAGTTCAG TTGGGTCTCA GCACTTCAAC 001120 001121 TCCTGGCTGG ATCCTGAGTT TTTCTTGGTT GCAGGGAAAG CTCTCACTGT CTGCATCTCC TTCCCCACCC CCACCTCCCG 001200 001201 TTTTGCAGCT GGCCATTCTG GCTGCCTCAC AGCTATCCTG CCTTAATTCA TATTTGATTG CCTGGACAGA ACCACCTAGA 001280 001281 ATGTCCAGAC GACTGCCACC CCTCAGCCAT GGGCCTGAGA TCCTGCCCTG ACCCCTTTCT TCAGACCCCA GGAGGACCCA 001360 001361 CAATCTCCCT GATCCCCACC TTCCCCAAAG TAGTCTCTAG GAAAAAAGCA GAGCTGGCTA AGGCCTGAGA CAGAGCCATT 001440 001441 TCTTCCCCTT GGGCAAACGA GGTAGACCCT CCCAGAGACC AATGCCGGAG AGGCCCGGAT CCGAGCTCCC TTTATATGGC 001520 001521 CTCCCCCATC ACGGATCCCG TGGGAAAACC TGACACACAA TGGCGCGGCT GCTCCGGGCA CCCTCAACCC AGCGCCCTGC 001600 001601 GGCAAGGCCT GGAGCAGCCT GGTGGTCTTG GTGCGCTCCC TTTGCCCTCC CCGCAACCCT CTGCGAGCAC TCGTCCCAGG 001680 001681 AGAGAGACCC TGGCTAGGGT GGAGGTTGCC TCCCGCAAAG GGCGCTGTTG CTGGGGCCTC CCCTCCCCCA TTCTCTTGTT 001760 001761 TTTCTGCTTC TGAGACTCGG GAGGGAGGCG GCAGGCGCCT CCGCAGCCGA TAGAGCAGCT ATTGTGACCT TCCCTTGCAG 001840 001841 AGCGGGAACC CAGCCGAGTG TGTACCGCCG GCGGTGGCGG GGGAGGAGGC GACTGTGTGT GCAAACCTCA GTCGGAGAGG 001920 001921 GGGTGGCGCT GGGAAATAAA GGTTTGCAAC CTCAGGCAGT TGGGCCCTTC AGAGGGCGTT TGGTTGTGTT TAGACTCGGT 002000 002001 TGCTTTCTAA CACTAATTGC AGGCTCAAAA GGAGCGGAAT TTATTGCGAA GGCCAAGGCG GCTTGTTTAT AATTTATGAA 002080 002081 GTTTTAAATG GCTGTGCCGC GGGCCCCAAC TTTCTCCTGG GCTGGCAGCC GCGCGGAAAC GCGCCCTCCC CCGGCTCGCC 002160 002161 GCCCACCCCC ACCCACGCCC CTCAGATGAG TCTCACGCCA ATGGCAGGAA CAGATCCGAG CTATTAGAAG TCTCGGGAAC 002240 002241 CTCCAGAGAT GACAGAAGGT GGCAGATTTG GTCCAAGCCC TCACCCCACT CGAGAGGAGG GCGTCGGGGA AAGCTGTGGC 002320 002321 TGCCTGGAGA GGGAGCCCCT GATCCCACAC TTCCTTGGAG CCTGAGTGTG AGAGGAAGCT AATGTCAGCC GGGCTAGACA 002400 002401 CCGTTTAAAG TCTAATCCAT GAAACCCTAA GCCGCAAATG ACACTTGGGG CTTTGCAGAC ACCCGCAGAC TCAGCAGATA 002480 002481 CTCCTGCGGT AACACACTGT GCTGGAAAAC GCCTGGAGCC ATGACACATT ACGTATTTTT ATTATTATTT ATGGTAGGAA 002560 002561 GAGGAGATGA GGGAAAATCG GGCTGAATCT AAGCAGCCTG ACAATTCAGC TAGTTAATAT TTAGCCAGTT AAGAAAGGTG 002640 002641 GTTGTAATCC CTGCAAGGCA GTGAGCTCTC CTTTGATTTC TCTAATAGCT TAAAAAATAA TTTCTTACAA GACAATTAGA 002720 002721 AATTCTGCCT ATGGATGCAA ATTTCATAGA AAACAATGTT AAGCCTTGAA AGTGGGTAAA ATGTGCCTTC GATTGAAGTC 002800 002801 ATTTATTCTG TTGATACACT TCTACAACAT ATATATTTTA AACCTGCTCA GATAAAGTGA AATTTAGCAA AAGGATATCC 002880 002881 CCTCCCACCC CCAAATCTAA CATGCCATTT TAAAAACAGC ACAAAGGAAC AAGGAAACTA AATATTATTT TGAGCCATCA 002960 002961 TTACTTAGTG AAATACAAAC ATCCAAATAG AAACGATTGT TGTTCCACCA GAAGATTAGA AGTTAAATTA TTTCCACACA 003040 003041 ATAGGAATTT CACTTCGTGT CATCGGGGAT GTAGTAGGAG TTGCATTATT TGTCAAACCA AATAGAGATG CGTGGGCTGA 003120 003121 CTTTGCTGAA ATACAAAAGA ATTGAACCTT ATCTGAAGGC GTTTTCTAAG TGGCCCCTCC CATAGAAGCC GGGAAGTTGT 003200 003201 CCACCCCTAG CAGAAGCCAC ATTAATATTT ACAAAATGGT TATAAATCCT TGAAGACATT TTTGCCCATT TTGTATAAGA 003280 003281 AACAGGAGTA AACAAGGTGT TTATACTTGT TATGTTTCAT TAATGAAATA TTATTACTTT ACATCTTGGG GAATGTTTGT 003360 003361 TTTCTTTAGA AATGAACTAA TGATCACTAC AAAGGAAGAA TTCATGACAG CATTTCCTAG AGAAAAAGCT TAGGTACGCG 003440 003441 TCTCCAAATG ATGGTTTCTT TTTCTACACC TTAGCTTTTT CATAGCTCAA TTCTTCTTAC AACTTCTCTT GTGTTTTTTC 003520 003521 TTTGATTGTC AGTTTTCTTT CAAAGTTTAT TTTTTGTTAC TTAATGCAGA AAAGTGAGCT CTGGTCTCCC CAGTGTAAAA 003600 003601 GTCGATTGTG TCTAGAAAGA AGGCAGTAAA AACAGCAAAG CAGGCGCTGC TTTCAAAAAG CCCTGCCCAA ATCATCCCGC 003680 003681 CCAGAACCTT CTTAGGTTTG CATCCTCATT TCCCCCTCCC CTAATTTCAA TAGCTGGAAA GTAATCAAGT ATCCCGAGAA 003760 003761 CCAATTTATG CACTAGACTC CAAATAAATA AAATATTTTT TTCAA |