NONHSAT076153
PCGEM1 (Prostate cancer gene expression marker 1) is a long non-coding RNA (lncRNA) overexpressed in prostate cancer (PCa) cells that promotes PCa initiation and progression and protects against chemotherapy-induced apoptosis [1].
Contents
Annotated Information
Name
PCGEM1: Prostate-specific transcript, LINC00071 (HGNC)
Characteristics
PCGEM1 is located at human chromosome 2q32.3 and spannining the genomic region of spanning the 27054 bp, consists of three exons that comprehends 1603 bp (GenBank) [2].
Function
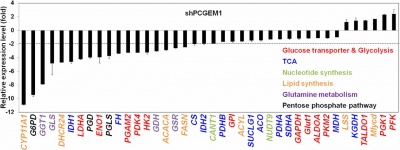
PCGEM1 regulates prostate cancer cell growth, tumor metabolism and proliferation of human synoviocytes [1][3][4][5].
PCGEM1 regulates metabolism at a transcriptional level that affects multiple metabolic pathways, including glucose and glutamine metabolism, the pentose phosphate pathway, nucleotide and fatty acid biosynthesis, and the tricarboxylic acid cycle. The PCGEM1-mediated gene regulation takes place in part through AR activation, but predominantly through c-Myc activation, regardless of hormone or AR status. PCGEM1 binds directly to target promoters, physically interacts with c-Myc, promotes chromatin recruitment of c-Myc, and enhances its transactivation activity [3].
Over-expression of PCGEM1 leads to inhibition of apoptosis induced by doxorubicin [6]. PCGEM1 and PRNCR1 interact with the androgen receptor (AR) bound at DNA-enhancer regions in a ligand-dependent fashion and facilitate the chromosomal looping between AR-bound enhancers and the promoter sequences of androgen-responsive genes [4].
Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells and nu/nu PCa tumor growth. Both downregulation of a tumor-promoting long noncoding RNA PCGEM1 or overexpression of the tumor suppressor miR-145 reduced the proliferation and invasive capacity of prostate cancer cells in vitro and in vivo [1].
PCGEM1 is involved in the hyperplasia of the synovial lining, a common characteristic of Osteoarthritic (OA). Exogenous overexpression of PCGEM1 inhibited apoptosis, induced autophagy, and stimulated the proliferation of human synoviocytes. The increased expression of PCGEM1 in human synoviocytes also suppressed the expression of miR-770. Transfection of the miR-770 precursor resulted in reduced proliferation, and induced apoptosis of human synoviocytes [5].
Expression
PCGEM1, was expressed exclusively in human prostate tissue, was dramatically upregulated in PCa tissues compared with normal prostate tissues. Prostate tissue-specific and prostate cancer-associated [2]. Cholesterols upregulate the expression of PCGEM1 even in androgen-insensitive prostate cancer cell lines while phytosterols reverse this effect [7]. PCGEM1 overexpression in LNCaP and in NIH3T3 cells promotes cell proliferation and a dramatic increase in colony formation, suggesting a biological role of PCGEM1 in cell growth regulation [8]. Overexpression of PCGEM1 stimulates proliferation andInhibits apoptosis of synoviocytes [5].
| Experiment | Forward primer | Reverse primer |
|---|---|---|
| Quantitative PCR | 5′-TGCCTCAGCCTCCCAAGTAAC-3′ | 5′-GGCCAAAATAAAACCAAACAT-3′[2] |
| siRNA | 5′-GCCCUACCUAUGAUUUCAUAU-3′ | 5′-AUAUGAAAUCAUAGGUAGGGC-3′[1] |
| qRT-PCR | 5′-CACGTGGAGGACTAAGGGTA-3′ | 5′-TTGCAACAAGGGCATTTCAG-3′[1] |
Regulation
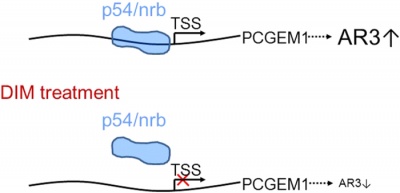
p54/nrb interact with the PCGEM1 promoter and regulate PCGEM1 [9].
Allelic Information and Variation
PCGEM1 polymorphisms may contribute to PCa risk in Chinese men. Men carrying single nucleotide polymorphisms (SNPs) of PCGEM1, i.e. rs6434568 AC and rs16834898 AC, had a lower PCa risk in comparison to the ones harboring CC and AA genotypes, respectively [10].
Disease
- Osteoarthritic
- Prostate cancer
Labs working on this lncRNA
- State Key Laboratory of Reproductive Medicine, Institute of Toxicology, Nanjing Medical University, Nanjing, China.[10]
- Department of Environmental Genomics, Jiangsu Key Laboratory of Cancer Biomarkers, Prevention and Treatment, Cancer Center, Nanjing Medical University, Nanjing, China.[10]
- Department of Genetic Toxicology, Key Laboratory of Modern Toxicology of Ministry of Education, School of Public Health, Nanjing Medical University, Nanjing, China.[10]
- Department of Urology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China.[10]
- Department of Environmental Genomics, School of Public Health, Nanjing Medical University, 818 East Tianyuan Road, Nanjing, Jiangsu 211166, China. [10]
- Department of Laboratory, Central Hospital of Panyu District, 8 Fuyu Dong Road, shiqiao, Guangzhou, Guangdong 511400, P R China.[1]
- The Second Affiliated Hospital of Guangzhou Medical University, Guangzhou 510620, China.[1]
- Department of Pharmacology and Toxicology, Cancer Institute, University of Mississippi Medical Center, Jackson, MS, USA.[9]
- Department of Radiation Oncology, University of Mississippi Medical Center, Jackson, MS, USA.[9]
- Department of Biochemistry, Cancer Institute, University of Mississippi Medical Center, Jackson, MS, USA.[9]
- Department of Radiation Oncology, Duke University Medical Center, Durham, NC, USA.[9]
- System Biosciences, Mountain View, CA, USA.[9]
- Department of Pulmonary Medicine, Tongji Hospital, Tongji University, Shanghai, China.[9]
- Department of Pathology, University of Mississippi Medical Center, Jackson, MS, USA.[9]
- College of Life Sciences, Zhejiang Sci-Tech University, Hangzhou, China.[9]
- Department of Biochemistry and Molecular Medicine, Comprehensive Cancer Center, University of California at Davis, Sacramento, CA 95817.[3]
- Institute of Molecular and Genomic Medicine, National Health Research Institutes, Zhunan Town, Miaoli County 350, Taiwan.[3]
- Department of Surgery, Center for Prostate Disease Research, Uniformed Services University of the Health Sciences, Bethesda, MD 20814-4799.[3]
- Department of Biological Sciences, College of Natural Sciences, Wonkwang University, Iksan, Chunbuk 570-749, Korea.[5]
- Department of Orthopedic Surgery, Wonkwang University School of Medicine, Iksan, Chunbuk 570-749, Korea.[5]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 He J-H, Zhang J-z, Han Z-P, Wang L, Lv YB & Li Y-G. Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells[J]. Journal of Experimental & Clinical Cancer Research. 2014, 33(1):72.
- ↑ 2.0 2.1 2.2 Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer[J]. Proceedings of the National Academy of Sciences. 2000, 97(22):12216-12221.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Hung C-L, Wang L-Y, Yu Y-L, Chen H-W, Srivastava S, Petrovics G et al. A long noncoding RNA connects c-Myc to tumor metabolism[J]. Proceedings of the National Academy of Sciences. 2014, 111(52):18697-18702.
- ↑ 4.0 4.1 Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T & Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer[J]. European Urology. 2014, 65(6):1140-1151.
- ↑ 5.0 5.1 5.2 5.3 5.4 Kang Y, Song J, Kim D, Ahn C, Park S, Chun CH et al. PCGEM1 stimulates proliferation of osteoarthritic synoviocytes by acting as a sponge for miR‐770[J]. Journal of Orthopaedic Research. 2016, 34(3):412-418.
- ↑ Fu X, Ravindranath L, Tran N, Petrovics G & Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1[J]. DNA and cell biology. 2006, 25(3):135-141.
- ↑ Ifere GO, Barr E, Equan A, Gordon K, Singh UP, Chaudhary J et al. Differential effects of cholesterol and phytosterols on cell proliferation, apoptosis and expression of a prostate specific gene in prostate cancer cell lines[J]. Cancer Detection and Prevention. 2009, 32(4):319-328.
- ↑ Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients[J]. Oncogene. 2004, 23(2):605.
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 9.7 9.8 9.9 Ho T-T, Huang J, Zhou N, Zhang Z, Koirala P, Zhou X et al. Regulation of PCGEM1 by p54/nrb in prostate cancer[J]. Sci Rep-Uk. 2016, 6:34529.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 Xue Y, Wang M, Kang M, Wang Q, Wu B, Chu H et al. Association between lncrna PCGEM1 polymorphisms and prostate cancer risk[J]. Prostate cancer and prostatic diseases. 2013, 16(2):139.
Sequence
>gi|64002|ref|NR_002769.1| Homo sapiens PCGEM1, prostate-specific transcript (PCGEM1), long non-coding RNA000081 tttaaagagc tatttatagg tccctggaca gcatcttttt tcaattaggc agcaaccttt ttgccctatg ccgtaacctg 000160
000161 tgtctgcaac ttcctctaat tgggaaatag ttaagcagat tcatagagct gaatgataaa attgtactac gagatgcact 000240
000241 gggactcaac gtgaccttat caagtgagca ggcttggtgc atttgacact tcatgatatc agccaaagtg gaactaaaaa 000320
000321 cagctcctgg aagaggacta tgacatcatc aggttgggag tctccaggga cagcggaccc tttggaaaag gactagaaag 000400
000401 tgtgaaatct attagtcttc gatatgaaat tctctgtctc tgtaaaagca tttcatattt acaagacaca ggcctactcc 000480
000481 tagggcagca aaaagtggca acaggcaagc agagggaaaa gagatcatga ggcatttcag agtgcactgt cttttcatat 000560
000561 atttctcaat gccgtatgtt tggttttatt ttggccaagc ataacaatct gctcaagaaa aaaaaatctg gagaaaacaa 000640
000641 aggtgccttt gccaatgtta tgtttctttt tgacaagccc tgagatttct gaggggaatt cacataaatg ggatcaggtc 000720
000721 attcatttac gttgtgtgca aatatgattt aaagatacaa cctttgcaga gagcatgctt tcctaagggt aggcacgtgg 000800
000801 aggactaagg gtaaagcatt cttcaagatc agttaatcaa gaaaggtgct ctttgcattc tgaaatgccc ttgttgcaaa 000880
000881 tattggttat attgattaaa tttacactta atggaaacaa cctttaactt acagatgaac aaacccacaa aagcaaaaaa 000960
000961 tcaaaagccc tacctatgat ttcatatttt ctgtgtaact ggattaaagg attcctgctt gcttttgggc ataaatgata 001040
001041 atggaatatt tccaggtatt gtttaaaatg agggcccatc tacaaattct tagcaatact ttggataatt ctaaaattca 001120
001121 gctggacatt gtctaattgt tttttatata catctttgct agaatttcaa attttaagta tgtgaattta gttaattagc 001200
001201 tgtgctgatc aattcaaaaa cattactttc ctaaatttta gactatgaag gtcataaatt caacaaatat atctacacat 001280
001281 acaattatag attgtttttc attataatgt cttcatctta acagaattgt ctttgtgatt gtttttagaa aactgagagt 001360
001361 tttaattcat aattacttga tcaaaaaatt gtgggaacaa tccagcatta attgtatgtg attgttttta tgtacataag 001440
001441 gagtcttaag cttggtgcct tgaagtcttt tgtacttagt cccatgttta aaattactac tttatatcta aagcatttat 001520
001521 gtttttcaat tcaatttaca tgatgctaat tatggcaatt ataacaaata ttaaagattt cgaaatagaa aaaaaaaaaa 001600
001601 aaa