

Genomic variation data and phenotypic data are core data in SorGSD. Furthermore, practical tools for sorghum research, such as gene ID conversion, homologue search and genome browser, are also provided here. The data resources and tools mentioned above can be obtained directly from the Homepage or by 'Variation', 'Phonetype', 'ID Conversion', 'Homologue Search' and 'Genome Browser' functions.
A total of 39,547,621 genomic variations (including 33,825,236 SNPs and 5,722,385 small INDELs ) of 289 sorghum accessions were provided in SorGSD. You can search the variations on the page of Variation Search.
On this page, users can use advanced operations below to search variations:
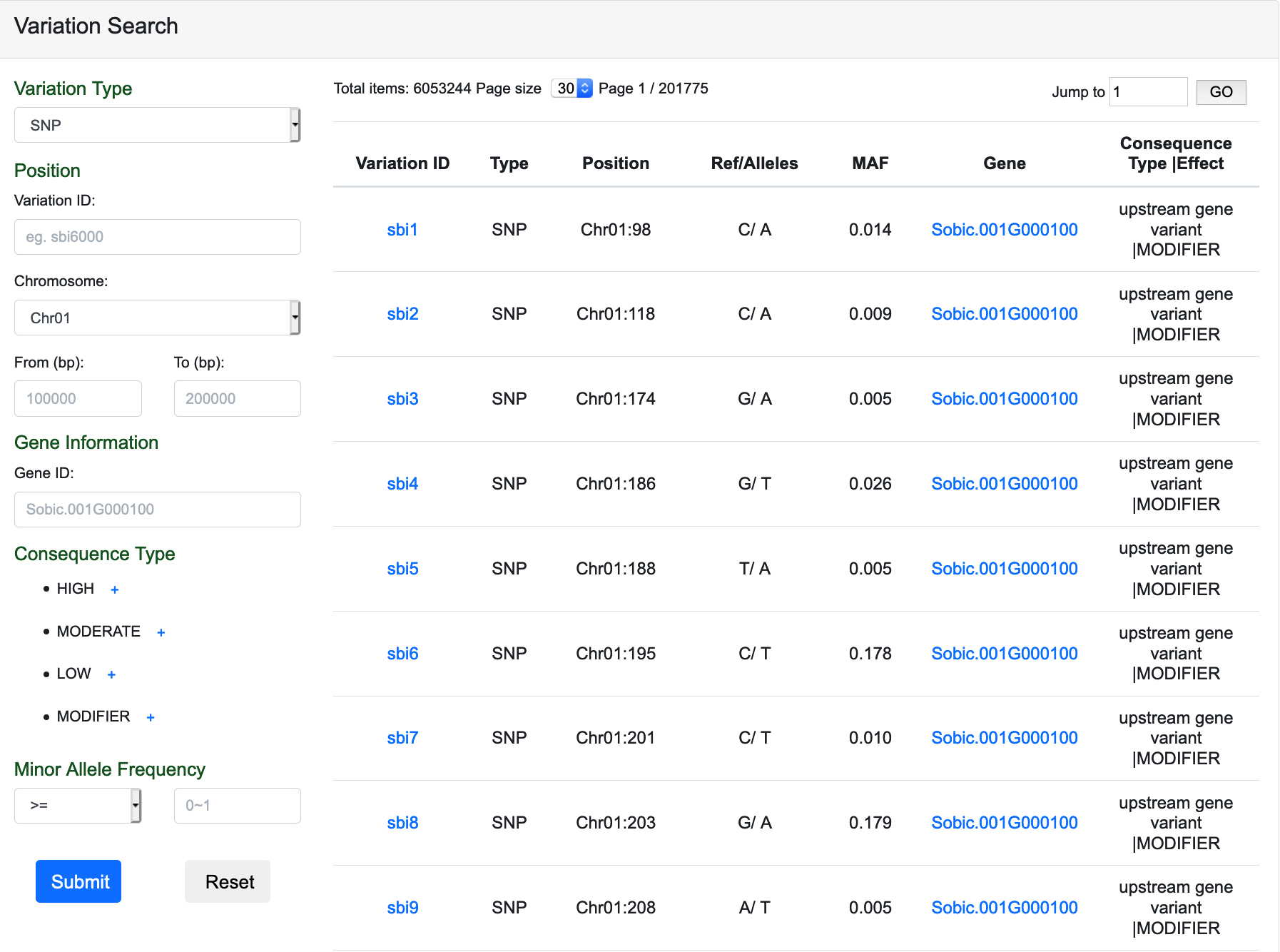
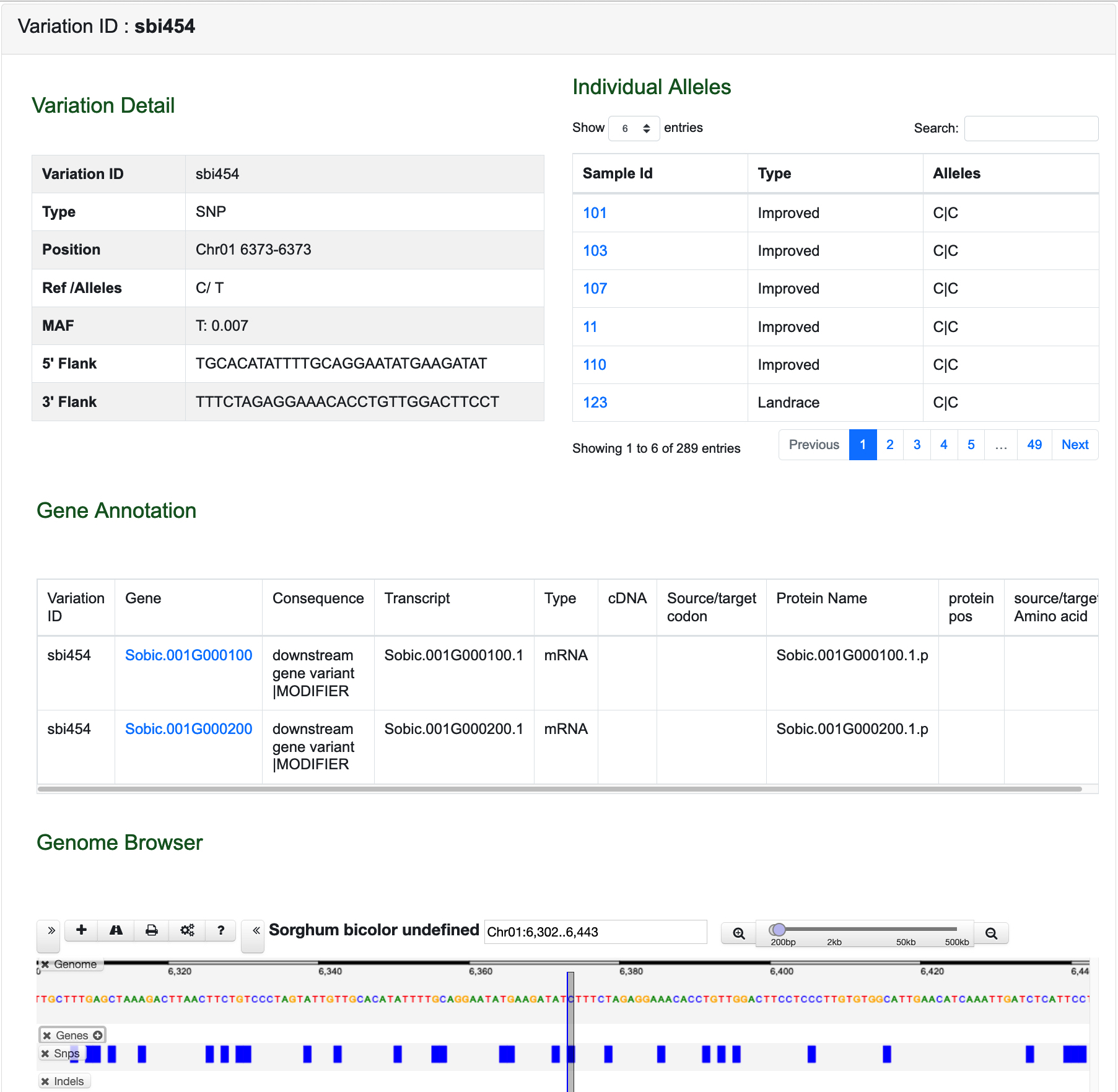
The link of Variation Information Page and Gene Information Page will also be given in this page.
The position of variation in genome will be displayed directly in Genome Browser.
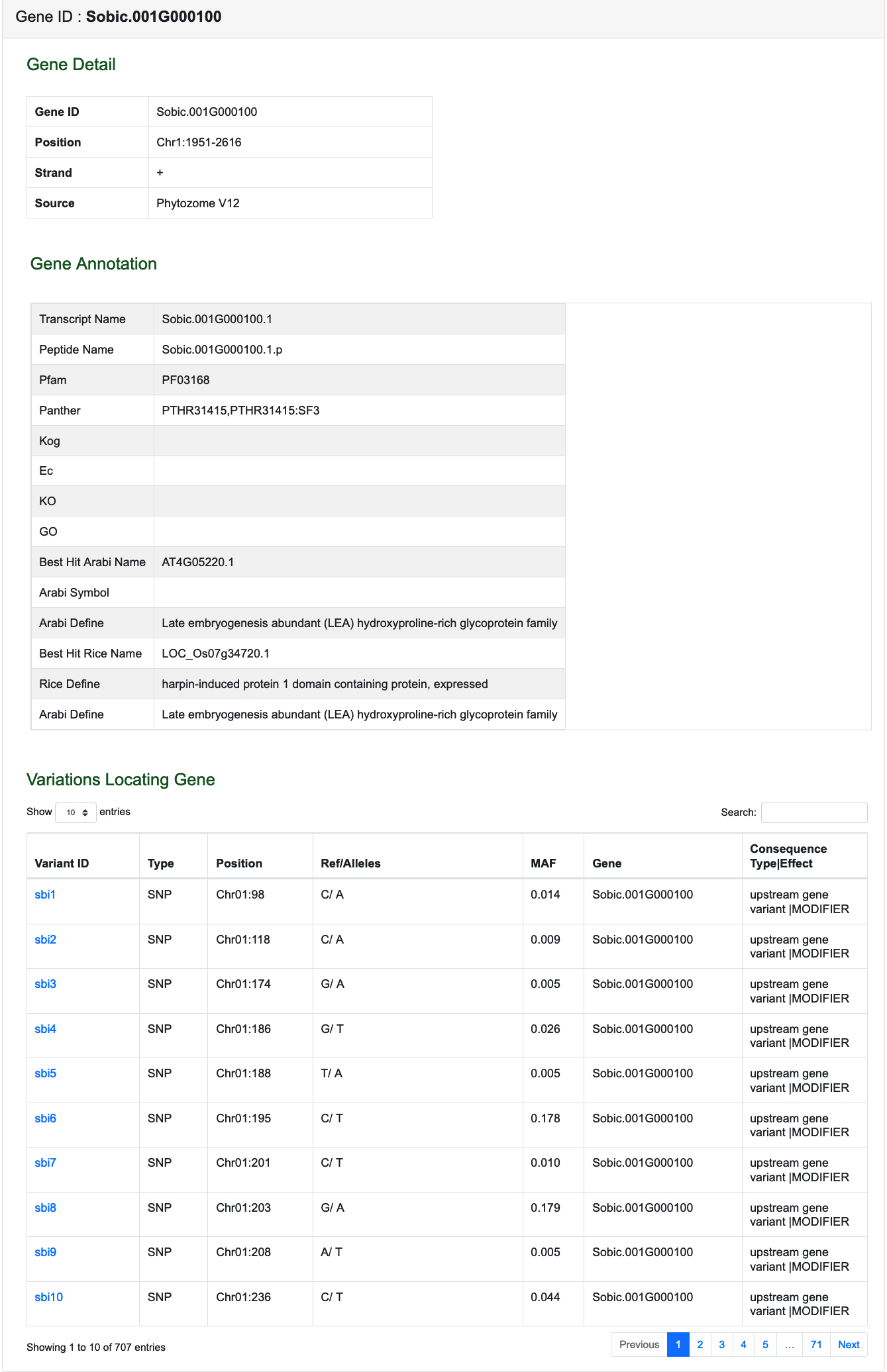
The Gene Information Page shows Gene Detail, Gene Annotation (include transcript and peptide name, annotations, best hit gene and define in Arabidopsis and rice) and Variations Locating Gene.
Phenotypic data of 289 accessions are provided in SorGSD. The pictures of panicles are taken in our laboratory and the detail information of each individual is collected from GRIN-Global. You can search the phenotype on the page of Phenotype Search.
On this page, users can browse the profile information (include Panicle picture, Sample ID, Plant ID, Plant Name, Origin, Taxonomy and Usage) of each individual. You can search any key information of individual by Search. Click the picture to see the high-resolution image. The accessions without panicle pictures are indicated with cartoons. Click the Sample ID to see the detail Information.

Here you can view the profile information of Phenotype Information and Phenotype Detail. Phenotype Detail includes Category, Descriptor, value and Study Environment.

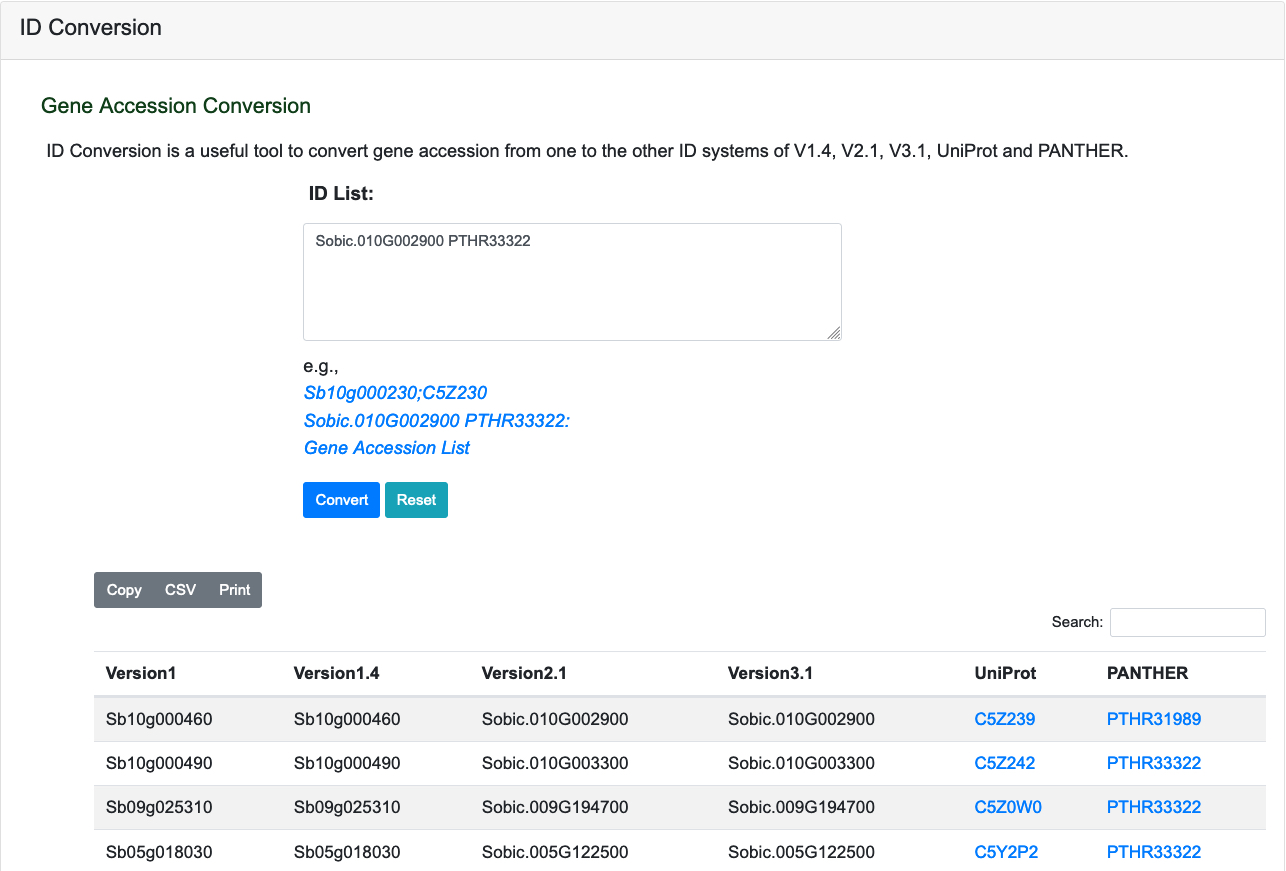
ID Conversion is a useful tool to convert gene accession from one to the other ID systems of V1.4, V2.1, V3.1, UniProt and PANTHER.
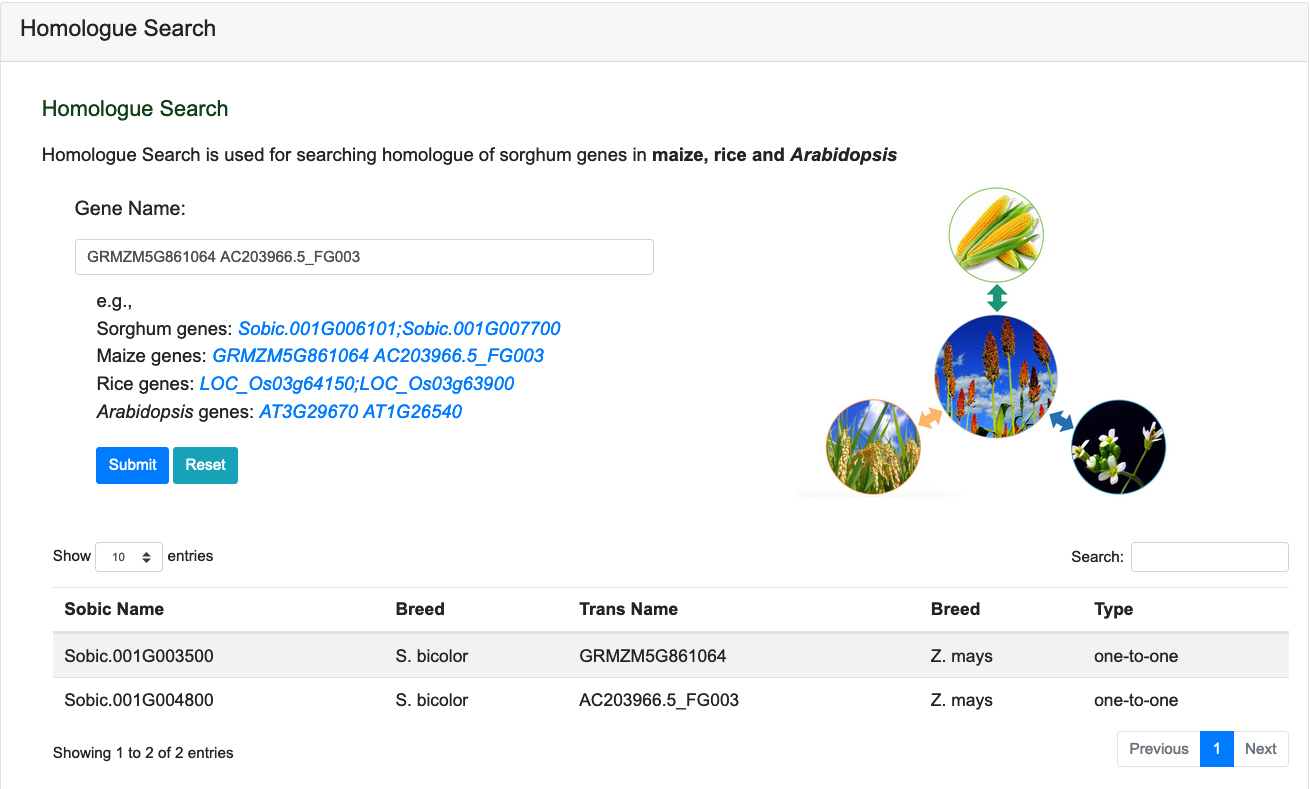
Homologue Search is used for searching homologue of sorghum genes in maize, rice and Arabidopsis.
SorGSD database uses Genome Browser to visualize several data types, including the variations, genes, transcripts and the density information of variations. Users would need to choose tracks and region of interest in the Select Tracks menu.
