Help
About
Database Commons is a manually curated catalog of worldwide biological databases, which has been frequently
updated and enriched since its inception in 2015. It aims to provide a full landscape of biological
databases throughout the world and enable easy retrieval and access to a specific collection of databases of
interest.
Database Commons integrates relevant information for all collected databases (including database name, URL,
description, hosted institution, related publication(s), contact information, etc.) and catalogues each
database based on its data type, species, subjects, locating, accordingly enabling people to easily find a
specific collection of databases of interest. We rank databases with total citations as well as the
normalized z-index to highlight popular and high-quality databases. Meanwhile, Database Commons allows
anyone to rate any database by considering data quality & quantity, content organization & presentation, and
system accessibility & reliability, facilitating efficient location of appropriate databases of interest.
Together, Database Commons features cataloguing databases under different criteria and incorporating
community rating on database utility, thus serving as a valuable resource for effective exploitation of all
publicly available databases.
Classification and Labelling
Databases are classified based on data type, data object and database subjects. In addition, major species
and keywords are tagged to further indicate the specific fields the database is related with.
Data Object
A database may encompass multiple data objects. In Database Commons, there are a total of 6 data
objects as
detailed below.
- Animal
- Plant
- Fungi
- Bacteria
- Archaea
- Virus
Data Type
A database may encompass multiple data types. In Database Commons, there are a total of 3 data types
as
detailed below.
- DNA: gene/chromosome/genome sequence, DNA mutation/modification, DNA structure,
DNA
elements including probe, primer, motif, repeat sequence, etc.
- RNA: RNA sequence, coding & non-coding transcripts, alternative splicing, RNA
editing/modification, RNA probe and primer, RNA motif and structure, RNA expression
- Protein: protein sequence, protein motif and domain, protein structure, protein
modification, protein-protein interaction, protein expression
Database Category
A database may encompass multiple database categories. In Database Commons, there are a total of 13
database
categories as detailed below.
- Raw bio-data: raw data of nucleic acid/protein sequencing and microarray, and
image, digit, video, audio from biological and medical research
- Gene, genome and annotation: gene/genetic element annotation, gene
structure/family/motif/domain annotation, genome annotation, comparative genome (metagenome,
pan-genome)
analysis and annotation
- Genotype, phenotype and variation: genotypes, phenotypes, multiple-scale
variations
(including SNP, INDEL, CNV, chromosomal rearrangement and other structural variation),
genotype-phenotype associations
- Phylogeny and homology: phylogeny reconstruction of genes/species, evolutionary
history/process/event among individuals/organisms, homology identification
- Expression: RNA/protein expression, expression abundance and pattern, RNA probe
or
primer used for gene expression detection, differential expression analysis
- Modification: DNA modification, post-transcriptional modification of mRNA and
non-coding RNA, post-translational modification of protein, modification
type/technology/function
- Structure: secondary, tertiary and quaternary structure of DNA/RNA/protein,
chromatin
structure
- Interaction: direct (physical) and indirect (functional) associations,
including
protein-protein interaction, RNA-protein interaction, DNA-protein interaction, gene regulatory
interaction, biochemical reaction, antigen and antibody, and genetic interaction
- Pathway: biological pathways for metabolic, signaling, gene regulatory analysis
- Health and medicine: disease variation/genotype-phenotype association, immune
reaction,
disease model, clinical biomarker, therapeutic target, drug & chemical compound,
pharmacogenomics and
pharmacodynamics, electronic health record
- Standard, ontology and nomenclature: standard, ontology and nomenclature for
biological
entities
- Literature: literature information, literature/text mining, textual annotation
based on
literature
- Metadata: metadata information for biological entities, e.g.,
project/sample/experiment/run/database/tool
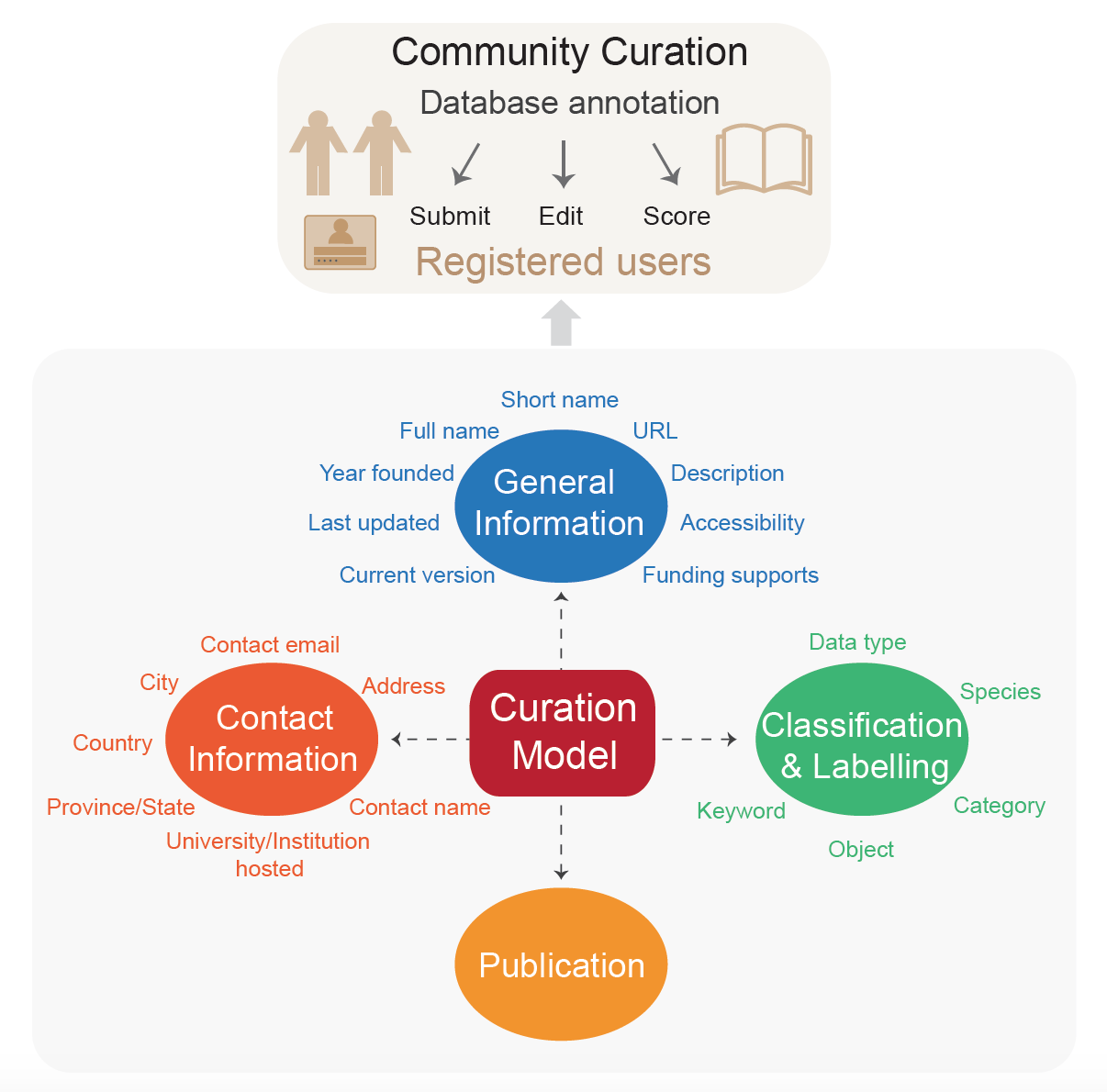
Curation Model
For each database, four sections are curated, including “General Information”, “Classification and
Labelling”, “Contact Information”, and “Publication”, involving 21 items. “General Information” details
basic information such as short name, full name, URL, availability. “Classification and Labelling” aims to
classify these databases based on their data type, data object and database subject, and lists more labels
to indicate the distinctive features of the database. “Contact information” is used to contact people who
take charge of database maintenance, and they are encouraged to participate in database curation. All the
information provided for each database entry is manually curated by multiple curators.
To ensure curation quality, only registered users are allowed to edit, submit, score database. We have
provided a curation handbook to introduce the standards and examples for each section and item, which is
available here.
Curation Permission
- To ensure content reliability, only registered users are allowed to edit/curate the database
information.
- Registered users can apply for curation permission by email. The Database Commons Team will
review the
applicants' qualification but does not guarantee the application will be approved.
- The Database Commons Team performs curation from time to time, and also encourages database
developers/curators/team-members to curate their own databases.
- Database Commons reserves the right to stop, to limit or to terminate your curation permission
for any
inappropriate or disruptive behavior on our website or revelant webpages.
- If you post or send offensive, inappropriate or objectionable content anywhere on or to our
websites or
otherwise engage in any disruptive behavior on any of our services, we may use your personal
information
from our security logs to stop such behavior and terminate your account. Where we reasonably
believe
that you are or may be in breach of any applicable laws we may use your personal information to
inform
relevant third parties about the content and your behavior.
Database Meta-Information
- In terms of accessibility, databases can be classified into alive and dead, where the former are
available, whereas the latter are unavailable temporarily or permanently due to various reasons.
- Database Commons collects not only active databases but also dead ones, just considering that
all
related meta-information of dead databases can also provide important history and insights for
users.
- For dead databases, their meta-information are obtained and extracted from their related
publications.
- The "Year Founded" indicates which year the database was founded. Albeit debatably, Database
Commons
considers the year of its first publication as "Year Founded".
- Many databases provide the last update information in the homepage and thus "Last Updated" was
curated based on this.
- URL is automatically obtained from publication and further manually curated by curators. This
URL
should direct users to the database homepage rather than other pages. If the URL has changed
during
update, it should be changed to the newest one.
- The "Accessibility" includes two options, namely, "Accessible", "Unaccessible", which are
manually
curated and checked by curators
- The "Description" is summarized by curators based on publication abstracts, which often should
be
concise and clear (1-3 sentences).
- Controlled vocabularies are used for three meta-information items, viz., "Data Type", "Data
Object",
and "Database Category".
- Three data types include "DNA", "RNA", and "Protein". A database may encompass multiple data
types.
If these three data types are not appliable, please select "Other".
- There are a total of 6 data objects, viz., "Animal", "Plant", "Fungi", "Bacteria", "Archaea",
and
"Virus". A database may encompass multiple data objects. If no species information is available,
input "NA".
- There are a total of 13 database categories. A database may encompass multiple database
categories.
- For "Species", Latin names of the organisms are required and should be selected from the
drop-down
box. If the database covers quite a large number of organisms, users could input names of the
major
organisms.
- Species list is obtained from NCBI Taxonomy database and some species may not be included in the
present list.
- Keywords are tagged to show the important features of databases. The singular form is preferred
rather than the plural form. All letters should be in lowercase.
- The contact information is provided to facilitate the update of database information, and it is
curated based on the contact details in the database or the related publications. To ensure
effective contact with database owners/developers, we give priority to the contact details shown
in
the database.
- For "University/Institution", official English full name of the university/institution is
required.
If the university has multiple campuses, the campus's name should be included, e.g., University
of
California Santa Cruz. If institutions are affiliated to an academy, the academy's name should
be
listed, e.g., Beijing Institute of Genomics, Chinese Academy of Sciences.
Database Citation & Age
- The "Citation" indicates the total citation count for a specific database, based on the summed
citations (indexed by Europe PMC) over all its related publications.
- Database age is calculated since the year of its first publication.
- z-index is calculated by dividing citation by database age, and this index is conductive to
reducing
influence of database age and enables relatively fair comparison between newly constructed
databases
and old well-established databases.
- Databases are ranked by z-index. Rank numbers among all databases and among specific database
category/categories are listed in the database page.
- For any given database, its related databases are classified into "Cited" and "Citing", where
"Cited" represents databases that cite this database, while "Citing" represents databases that
have
been cited by this database.
- Curation events are recorded by day. Curators may curate a specific database for many times per
day,
but this would be registered as one record in "Record metadata".
Evaluation System
The popular and high-quality biological and biomedical data, contributes greatly to
biological and biomedical discoveries. Therefore, we incorporate evaluation system in Database Commons to
measure database quality and impact.
There are four rating items, “Citation”, “z-index”, “Accessibility”, and “Community reviews”. “Citation” of
a certain database is the total citations (indexed by Europe PMC) of all its published papers, and high
citations always indicate popular and high-quality databases. “z-index” is calculated by dividing total
citations by database age, and this rating item is conductive to reducing influence of database age and
enables relatively fair comparison between newly constructed databases and the old well-known databases.
“Accessibility” represents the accessibility status of the homepage, including the manually curated status
and the analysis of HTTP status codes (listed as follows). “Community reviews” requires community engagement
and it is a comprehensive evaluation of data quantity and quality, content organization & presentation, and
system accessibility & reliability. Among the four rating items, Citation and z-index have been
automatically calculated for all biological databases, and users can rank databases and refine search
results based on the two items.
Community Rating
Database Commons features community rating on database utility by taking account of the following
three
criteria.
Data quality & quantity: consider data integrity, accuracy, standardization, consistency
and
comprehensiveness
Content organization & presentation: consider whether content is organized in an
appropriate
manner which makes content easily readable and understandable and is presented by user friendly
web
interface
System accessibility & reliability: consider whether system is always accessible and
reliably
working
A database containing high-quality curated data is abortive if data is poorly organized or
presented.
A database containing high-quality curated data is unavailing if this database cannot be accessible
or
reliably working.
HTTP Status Codes
Here is a list of HTTP status codes with a brief explanation, which are represented by three digits
and fall
into two classes.
Accessible
2xx Success: e.g., 200 OK, that is standard response for successful HTTP
requests.
3xx Redirection: e.g., 301 Moved Permanently
Unaccessible
4xx Client Error: e.g., 403 Forbidden, 404 Not Found
5xx Server Error: e.g., 500 Internal Server Error, 503 Service
Unavailable
More information about HTTP status code can be found at Wikipedia.
In addition, unexpected exceptions including timeout, errors occurred when sending requests, etc.,
are
indicated by "-1".
Database Usage
1. How to browse the biological databases?
In the browse page, all users can browse the biological databases by
‘Country/Region’, ‘Institution (Top 30)’, ‘Database Category’, ‘Data type’ or ‘Data object’ by selecting
specific category from the drop-down boxes on the left of the page. Also, it is easy to view the databases
by ‘z-index’, ‘Citation’, ‘Short name’ and the ‘Founded year’.
2. How to search the biological databases?
The home page provides global search for name, category, country, data type, etc. Search page allows both
global search and advanced search, where users could quickly retrieve a specific group of databases of
interest with customized filters.
3. How to submit the biological databases?
Only registered users are allowed to submit new databases in Database Commons. Please email us first if you
would like to take part in the curation work. Curators will be given basic training for database curation,
classification, and usage of the curation platform. You are able to curate or edit after administrator have
upgraded the privileges.
After login, click on ‘Submit’, and then input the database information for four sections based on the
structured curation model. The curation handbook details the curation rules for each item.
4. How to edit the biological databases?
To ensure curation quality, only registered users are allowed to edit. Users can edit the databases by
clicking the button nearby the database name in the database page, and update the information in the
curation page. Don’t forget to click on ‘Save’ when you have made any changes.
5. How to score the biological databases?
To ensure curation quality, only registered users are allowed to score. Users can select the star number of
‘Data quality & quantity’, ‘Content organization & presentation’ and ‘System accessibility & reliability’,
respectively, and then click on ‘Submit a review’.
More stars indicate higher quality.
6. How to cite Database Commons?
Database Commons: a curated catalogue of worldwide biological databases (in preparation)
Related Publications
Database Resources of the National Genomics Data Center, China National Center for
Bioinformation in 2022. Nucleic Acids Res, 2022. 50(D1): p. D27-D38.
[PMID=34718731]
Database Resources of the National Genomics Data Center, China National Center for
Bioinformation in 2021. Nucleic Acids Res, 2021. 49(D1): p. D18-D28.
[PMID=33175170]Database Resources of the National Genomics Data Center in 2020. Nucleic Acids Res, 2020.
48(D1): p. D24-D33.
[PMID=31702008]
Database Resources of the BIG Data Center in 2019. Nucleic Acids Res, 2019. 47(D1): p. D8-D14.
[PMID=30365034]
Contact Information
National Genomics Data Center,
Beijing Institute of Genomics,
Chinese Academy of Sciences and China National Center for Bioinformation,
Beijing 100101, China
Email: databasecommons(AT)big.ac.cn
Tel: +86 (10) 84097845

