| EpiCancer |
|---|
| Epitranscriptomics in Cancers |
1: Introduction
1.1 What is m6A?
N6-methyladenosine (m6A) is one of the most abundant epitranscriptomic marks on eukaryotic mRNA, and it mainly occurs at the consensus sequence motif of ‘RRACH’ (R=purine, H=A, C or U). It is written by the methyltransferase complexes (including METTL3, METTL14, WTAP, and KIAA1429, etc) and erased by demethylases (FTO and ALKBH5) in mammals. This process is dynamically reversible, and the RNA with m6A modification will be recognized by the m6A binding proteins (including YTH domain family, IGFBP family, HNRNPA2B1, etc.) and perform functions, such as mRNA translation, splicing, nuclear export, and RNA degradation.

Figure 1. The molecular mechanism of m6A
Note: RNA N6-methyladenosine modification in cancers: current status and perspectives. Deng X et al. 2018.Cell Res.
1.2 The importance of m6A regulators in cancer
Recently, the research between m6A RNA methylation and cancers has received extensive attention. More and more evidence shows that m6A RNA methylation is closely related to the occurrence and development of many cancers. And of course, m6A regulators (methyltransferase complex, demethylase and recognition proteins) are also important regulators in the occurrence and development of cancers, cause their expression level can directly affect the level of m6A modification, which in turn determines the pathological process of cancers. Therefore, studying the biological regulation process of m6A regulators in different cancers is of great significance for understanding the pathogenesis of cancer and screening anti-cancer drugs.
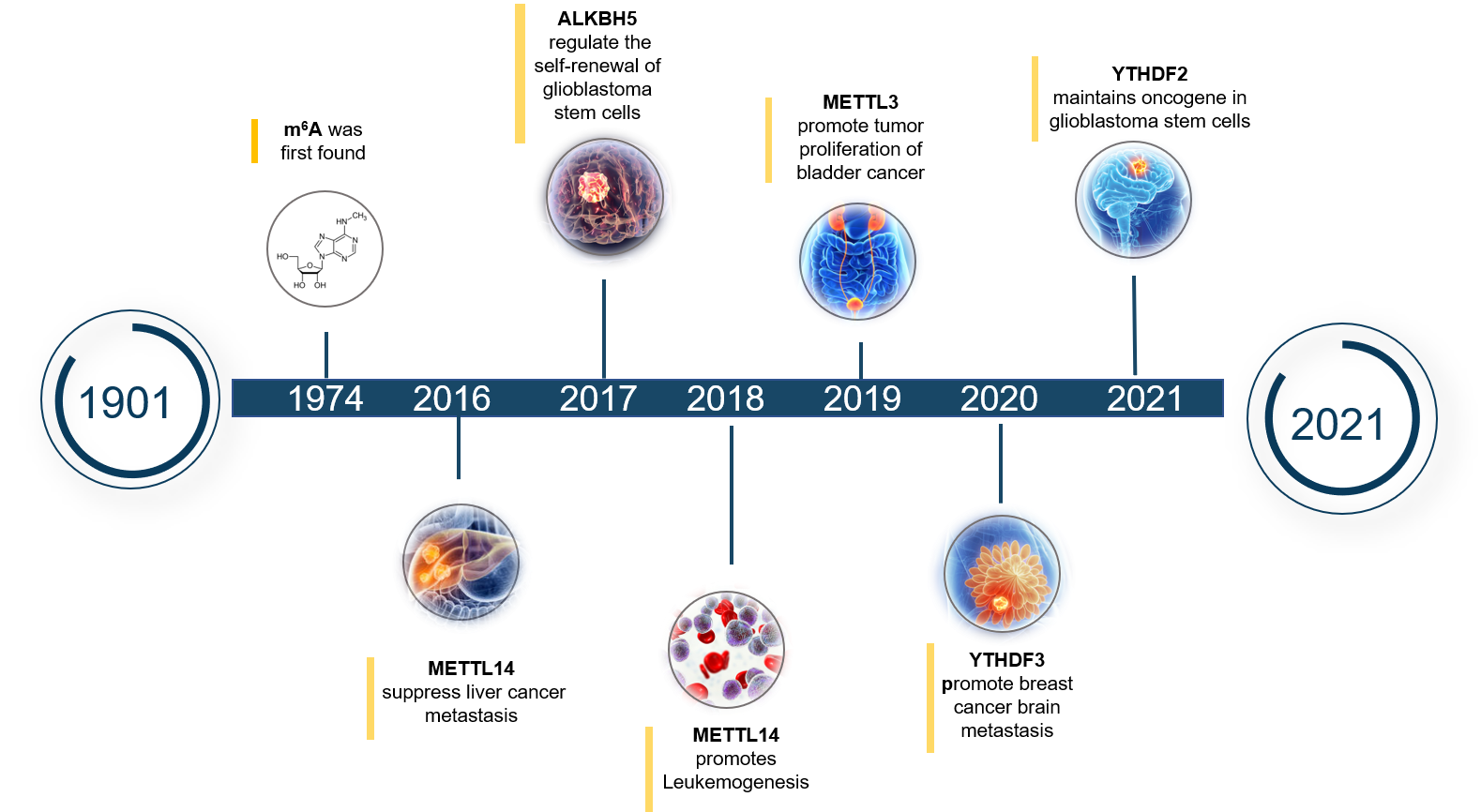
Figure 2. The research progress of m6A regulators in cancers
1.3 The regulation process of m6A in cancers
As the picture shows, m6A regulators can alter the m6A of the downstream target, thus influence the tumor progression, proliferation, and so on. So the upstream factors like DNA methylation and miRNA, which can regulate the expression of the m6A regulator genes, are also key effectors for the m6A-related cancers. Besides, some miRNA, RBP, RNA editing enzyme, and splicing factors which may compete for the m6A binding site to influence the m6A level, are also important effectors for m6A-related cancers. So in our database, not only we have the curated knowledge from the articles, but also we have the predicted upstream factors which may influence the m6A regulator genes to regulate the cancers and the predicted potential factors which may compete for the m6A binding site to influence the cancer process.
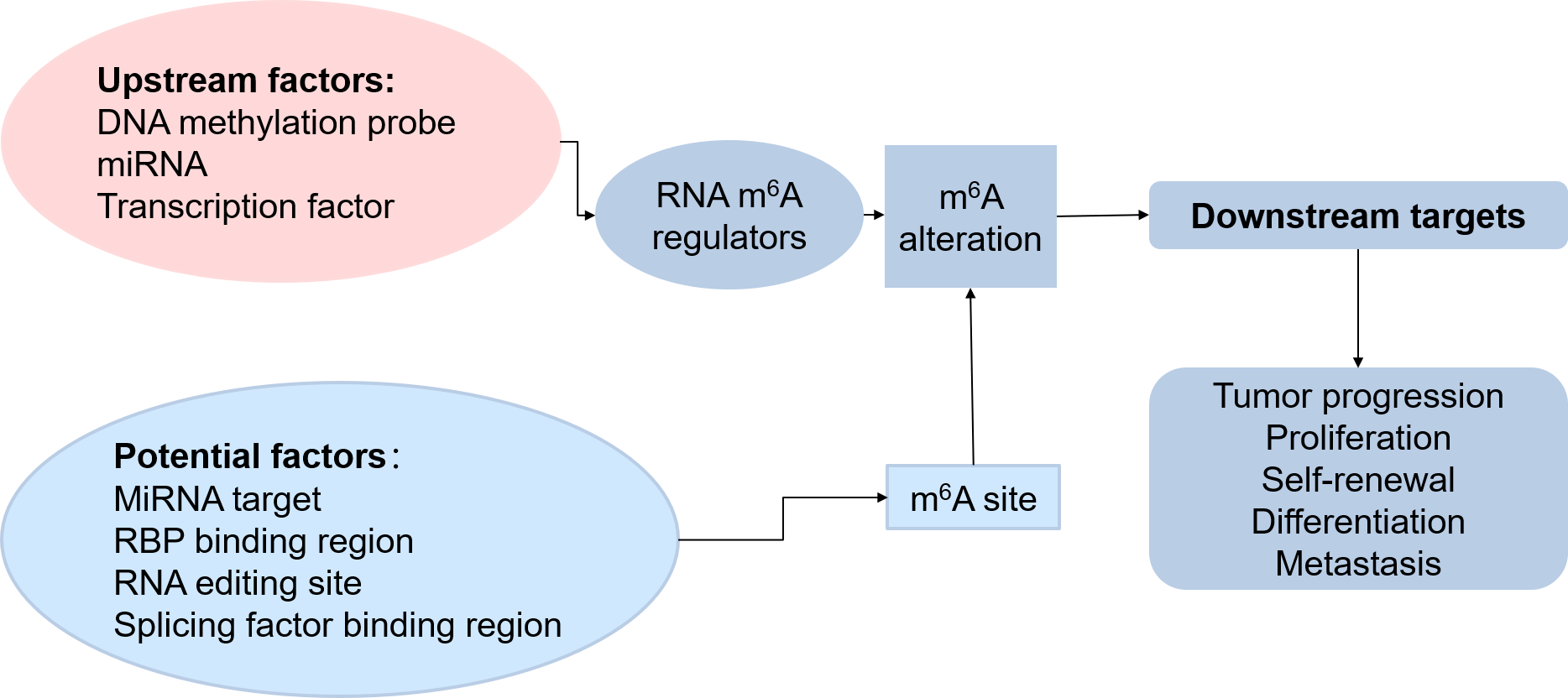
Figure 3. The regulation process of m6A in cancers
2. Datasets
The current version of EpiCancer incorporates 40 cancers,15 m6A regulators, and 11 m6A-regulator-related drugs curated from 139 publications. The database also incorporates millions of m6A sites analyzed from the curated m6A data; 100 analyzed DNA methylation probes, 514 miRNAs and 591 transcription factors which may influence the m6A regulator genes; and millions of miRNA, RBP, RNA editing enzyme, and splicing factors which may compete for the m6A binding sites.
3. Data Structure and data source
3.1 Curated knowledge
Articles from PubMed
3.2 Data
| Data | Source | URL |
|---|
4. Pipeline construction
Here is how EpiCancer organized.

5. User guide
5.1 Getting Started
We present EpiCancer, a curated knowledgebase of epitranscriptomic m6A regulators (also called WERs) in cancers that integrates (1) functional regulation in cancers, (2) drugs targeting WERs, (3) regulation network covering miRNA/DNA methylation/transcription factor, (4) and multi-dimensional competitive binding or targeting sites/regions.
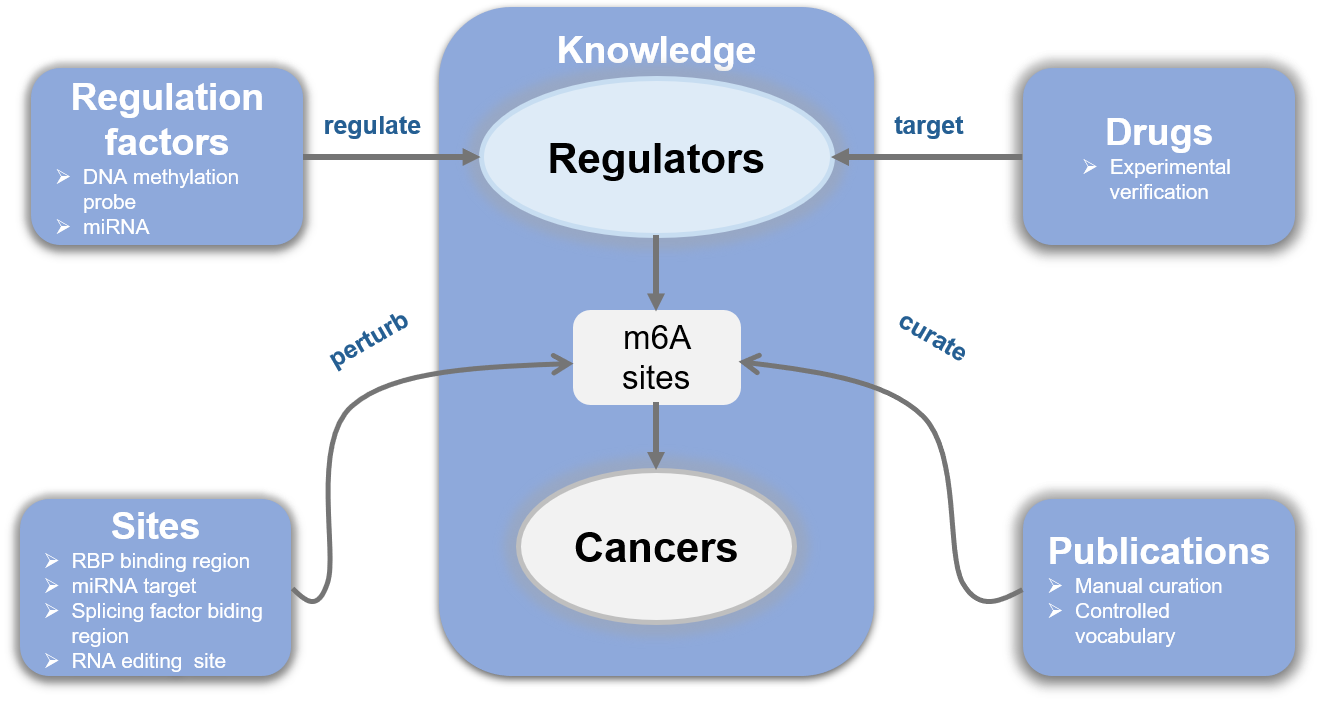
For convenience, we provide a user-friendly web interface for EpiCancer database. On the Home Page, the Quick search feature allows users to query data at three different levels, including “Regulator”, “Cancer type”, and “Gene symbol”. The schematic diagram demonstrates how our database is built. And the lowest heatmap is a general exhibition of the WERs’ functional regulation numbers in different cancers. Both the regulators and the cancers are clickable and will take the users to the same website where search button would guide to, which is explained in detail below.

5.2 Browse
5.2.1 m6A regulators
This page displays (1) a functional regulation information table of 15 regulators in 40 cancers, (2) a simple stat of all regulators’ publications in different cancers.
- In the table part, you can use the filtering options to refine your results. The following are the Illustration for filter types and other headers:
Regulator: 15 regulators in the present version.
Cancer: 40 cancers in the present version.
Role: The role of m6A regulators in cancers, including oncogene and suppressor.
Function: Cancer processes the regulator effect: including tumorigenesis, proliferation, apoptosis, self-renewal, stability, progression, transformation, invasion, migration, and metastasis, resistance.
Upstream: The experimentally validated upstream factors which can regulate the m6A regulators during cancer process.
Downstream: The experimentally validated downstream targets of m6A regulators.
Details: A simple description of the biological process.
Publication: Click the title, then you will jump to a new page for single publication. This page displays (1) The basic info of this publication, including PubMed ID, Accession, Abstract, and so on. (2) All the curated regulation knowledge of this article. (3) If this article has sequencing data, a basic stat and top 200 significant modified peaks is shown. PMID: External link to PubMed.
- In the profile part, you can click the m6A regulators on the y axis directly, then you will jump to a new page and get (1) the basic info of the m6A regulator gene, (2) all curated functional regulation info about this m6A regulator, (3) all the upstream regulation factors (miRNAs, DNA methylation probes, and transcription factors) of this m6A regulator, (4) and all drugs targeting this m6A regulator.


5.2.2 Cancers
This page provides stat of knowledge of regulation information in cancers, and gene ontology enrichment of m6A modified genes in cancers.
In the first profile part, you can click the cancers on the y axis directly, then you will jump to a new page and get (1) a stat of publication numbers of this cancer in all m6A regulators, (2) number of modified peaks and genes in this cancer type, (3) all curated functional regulation info about this cancer type, (4) all peaks identified in this cancer type if there are any datasets in this cancer type.
In the second part, the table is similar to the one in "Browse-m6A regulators part".
The third part displays a Gene ontology of all genes which are modified by m6A (left), and Gene ontology of m6A genes that are modified in more than 3 samples (right).

5.2.3 Sites
EpiCancer provides users an interactive genome browser to browse the methylation sites in different gene regions.
By enter an interval and click search, you will get an atlas of m6A modification sites, miRNA-targets, RNA-binding protein(RBP) binding regions, RNA editing sites, and splicing factors binding regions curated from other resources.
5.2.4 Regulation network
Regulation network in pan-cancer level is built based on m6A regulator genes and their upstream regulation factors (miRNA and DNA methylation transcription factor).
By choosing one or more m6A regulators, you will get an upstream regulation network of them, and the detailed information is shown in the tables below.
5.2.5 Drugs
This page provides the targeted drugs of m6A regulators in cancers. The overall information is shown in the sunburst picture and detailed regulation information is shown in the table below.
By choosing an m6A regulator, you will get all the targeted drugs on the table.
5.2.6 Publications
This page provides all curated publications with the basic information of the article and regulators or cancers mentioned in each article.
5.3 Statistics
All figures here are clickable via the text on axis except the "Number of publications" and "Data types" pics.