ParaAT (Parallel Alignment and back-Translation, is a parallel tool that parallelly constructs protein-coding DNA alignments for a large number of homologs. ParaAT is well suited for large-scale data analysis in the high-throughput era, providing good scalability and exhibiting high parallel efficiency for computationally demanding tasks.
The resulting alignments can be outputted into different formats, including fasta, axt (for KaKs_Calculator), paml (for PAML), codon and clustal. Documentation and usage information can be found here.
The answers to Frequently Asked Questions are summarized.
Please send bugs or advice to Zhang Zhang (zhangzhang.cn at gmail dot com or zhangzhang at big dot ac dot cn) or visit the Zhang Lab of Computational Biology and Bioinformatics in Beijing Institute of Genomics, Chinese Academy of Sciences.
Zhang, Z., Xiao, J., Wu, J., Zhang, H., Liu, G., Wang, X. and Dai, L. (2012) ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments, Biochem Biophys Res Commun, 419(4):779-7
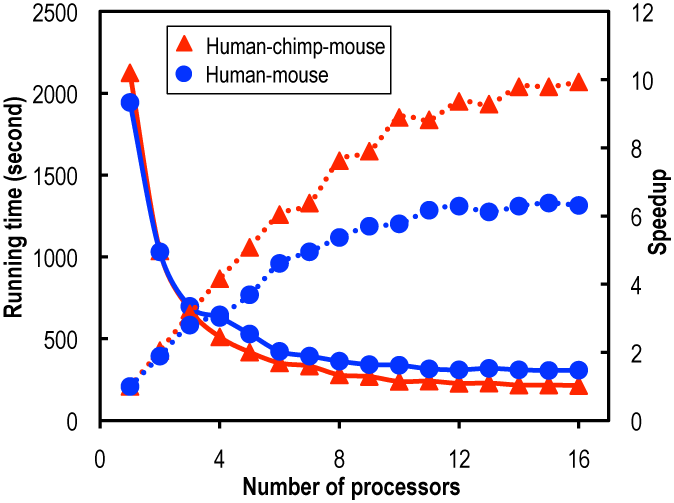
Speedup (dotted lines) and running time (solid lines) for constructing protein-coding DNA alignments using 1–16 CPUs. Two datasets were used: 15,430 human–mouse homologous gene pairs (circles) and 9712 human–chimp–mouse homologous gene trios (triangles). ClustalW was employed for aligning protein sequences.