About CIRIhub
CIRIhub is established as a automatical analysis platform to help researchers achieve biological analysis for circRNAs based on RNA-seq or Microarray datasets.In the first step, we recommed CIRIquant software to idetify back-splicing sites and expression of genes from RNA-seq dataset. CIRIhub will analyze a pre-published stomach tumor total RNA-seq dataset, which contains circRNA expression and gene expression of tumor and normal tissues.
Module-"One Click"
One-stop analysis for one circRNA, a list of circRNAs or in-house RNA-seq data.

"One-click" Input for in-house RNA-seq dataset
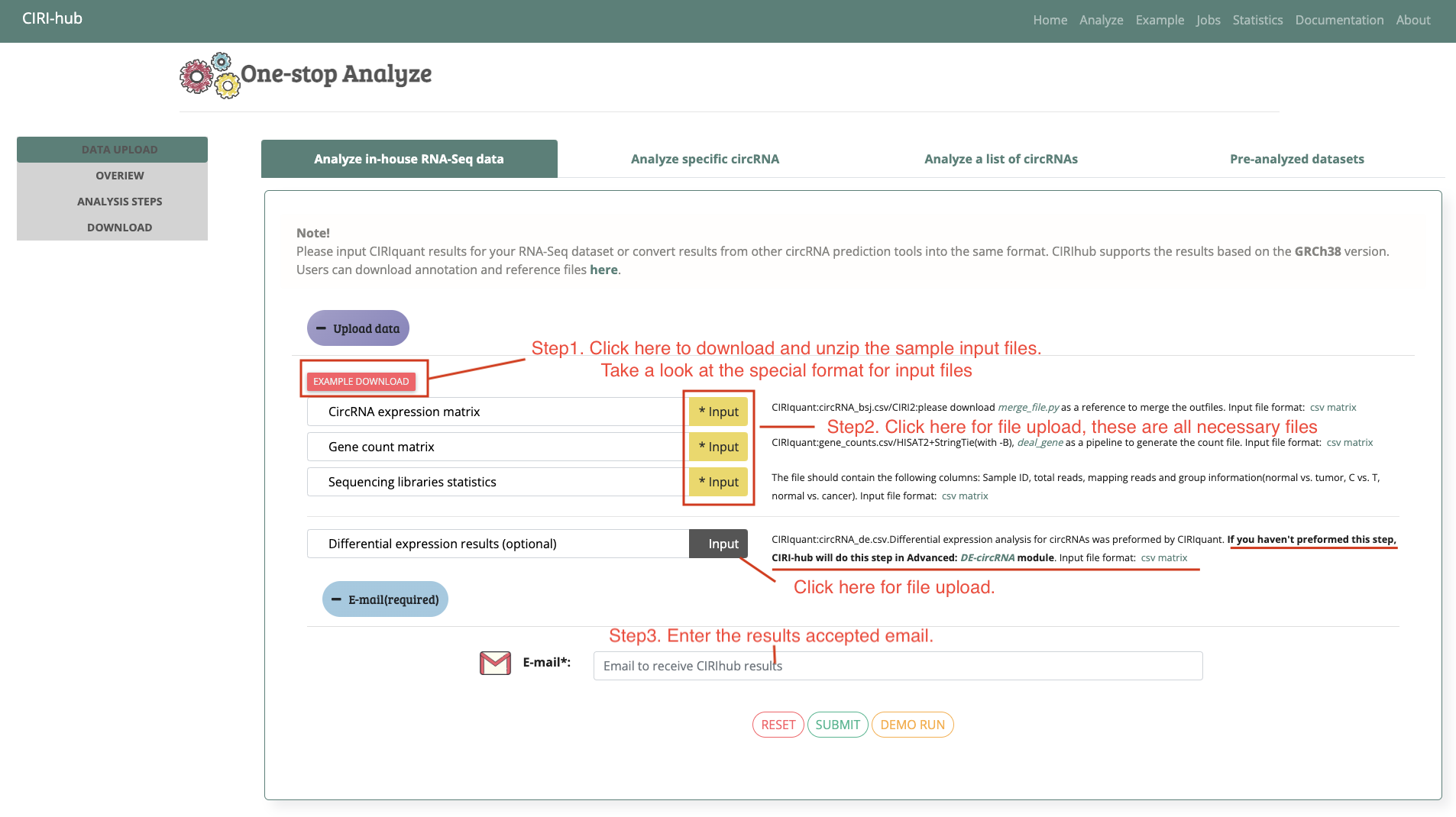
"One-click" Running and Output(Online)

"One-click" Output(Email)

"One-click" Output(Browse details)

"One-click" Output(Customize sub-figure)

"One-click" Input for specific circRNAs(same as a list of circRNAs)

Module-"Advanced"
Run specific analytical steps with customized parameters.

"Advanced" Input for DE-circRNA

"Advanced" Output

CIRIhub Manual(PDF)
CIRIhub Analysis Description
 Sample clustering analyses
Sample clustering analyses
Clustering of samples based on gene and circRNA expression. Four clustering methods:PCA, UMAP,MDS,TSNE are available for screening. One click result displays UMAP, and other clustering results are displayed in the details of the sample clustering module.
 circRNA filtering
circRNA filtering
circRNAs are filtered by the criteria: detected more than 10% sample. You can click the detail circRNA filter module to customise the cut-off.
 circRNA types
circRNA types
circRNAs are classified into 6 types: exon,intron,intergenic,antisense, 5UTR,3UTR.
 Reported circRNAs
Reported circRNAs
The number of circRNAs that have been reported in public normal RNA-seq data (circAtlas) and tumour datasets (MiOncoCirc & CSCD2).
 Conversation score
Conversation score
circRNAs are ranked by the multiple conserced score(MCS) from circAltas. Conservation analysis is always used to screen the potential functional circRNAs, relatively high conversation score represents potential importance of circRNAs.
 Multiple tissue expression
Multiple tissue expression
Apply circRNA expression analysis in normal tissues and cancer types, significance tests are performed in paired normal and cancer datasets.
 Up-regulated and down-regulated across cancer tissues
Up-regulated and down-regulated across cancer tissues
Fold change of circRNAs and genes across multiple cancer tissues.
 DecircRNA(Differential expression)analysis
DecircRNA(Differential expression)analysis
Differential circRNA expression in the uploaded dataset and interested dataset(both normal and cancer datasets).
 Correlation analysis
Correlation analysis
The worse correlation between circRNA and host gene were analysed more than once in previous studies. Prediction and analysis of the associated gene is the key step to analyse circRNA performance.
 GO and KEGG enrichment
GO and KEGG enrichment
GO and KEGG enrichment analysis of significant up- and down-regulated circRNAs.
 Without Job ID
Without Job ID
Users can analysis the expression, conservation, rbp, micorRNA sponge of circRNA which reported in circAltas and Mioncocirc datasets.
 With Job ID
With Job ID
Samples were grouped into high and low group based on the expression of interested circRNA. Differetial gene expression of high and low group were performed and then GSEA analysis were preformed.